1000/1000
Hot
Most Recent

There is still a lack of reliable and specific markers for the detection and staging of thyroid cancer. Fine needle aspiration biopsy is the current diagnostic gold standard but indeterminate results or an inability to discriminate different carcinomas, requires additional surgical procedures to obtain a final diagnosis. Metabolomics has the potential to identify molecular markers of thyroid cancer and identify novel metabolic profiles of the disease, which can, in turn, help in the classification of pathological conditions and lead to a more personalised therapy, assisting in the diagnosis and in the prediction of cancer behaviour. This review considers the current results in thyroid cancer biomarker research with a focus on metabolomics.
Compared to proteomics and transcriptomics, thyroid cancer metabolomic studies have featured in relatively few papers over the last ten years. However, recent technical improvements in both hardware (mass spectrometers with higher mass accuracy, SWATH data-independent MS acquisition and ion-mobility MS) and software (improvement of metabolite identification databases, as well as metabolite biological integration and NMR automatic identification and quantification) have allowed metabolomics to emerge as a standalone method for profiling of thyroid cancer samples [1][2][3]. Although this review is focused on NMR and MS metabolomics, it is worth mentioning that other techniques could also be applied. Raman spectroscopy also has an interesting diagnostic potential: by analysing the vibrational modes of chemical bonds, it can identify non-specific molecules, such as proteins, lipids or nucleic acids, that may just be enough to distinguish between malignant and benign samples [4][5].
One of the first metabolomic studies that attempted to address the lack of diagnostic power in thyroid cancer was in 1994 and consisted of a 1H NMR study of 19 malignant and 24 benign patient tissue samples (Table 1). The authors were able to identify triglycerides and lysine as potential discriminatory metabolites, but the method’s specificity was only 52% [6]. Two years later, the same authors applied two-dimensional NMR spectroscopy, which improved the resolution of metabolite signals, allowing a higher number of metabolites to be monitored. However, this only led to a moderate improvement in the method specificity [7].
By the beginning of the 21st century, NMR spectroscopy had emerged as the main technique for performing metabolomic analysis. The first proof-of-concept studies in thyroid cancer used either magnetic resonance spectroscopy imaging (MRSI) [8][9][10] or 1H NMR spectroscopy on excised tissue samples or deproteinised tissue extracts [11][12]. One interesting feature of the study of King et al. is that it was one of the first studies to identify choline as a metabolite whose levels were changed in thyroid cancer [8]. This was confirmed in subsequent studies and choline has since often been proposed as a thyroid cancer biomarker. However, it should be emphasised that, while MRSI is non-invasive, the standard 1.5T systems in current clinical use are limited to detecting a handful of highly abundant metabolites, such as choline, within relatively large voxel volumes (≥1 mL) [8][9][10].
The first high-resolution magic angle spinning (HR-MAS) NMR metabolomics study [13] and the first MS study [14] that we found in our literature search were both published in 2011. HR-MAS allows spectra to be obtained from intact biopsy samples of 10–40 μL volumes with signal resolution approaching that of high-resolution NMR spectra of tissue extracts. The study of Jordanet al., although using a limited number of samples, had the benefit of being able to compare results of tissues with those of aspirates. The study of Yao et al. analysed the serum of 30 papillary thyroid carcinomas (malignant), 80 nodular goitres (benign) and 30 healthy controls and found that malignant and benign samples were correlated with changes in lipid metabolism, with 3-hydroxybutyric acid, an intermediate product of fatty acid metabolism, particularly important. One year later, the group of Prof. Caldarelli published two similar papers [15][16] using HR-MAS NMR on tissue samples. These revealed increased phenylalanine, taurine and lactate levels, and a decrease in choline and choline derivatives and myo- and scyllo-inositol levels in malignant tissues compared to benign. However, when these data were modelled using orthogonalised partial least-squares discriminant analysis (OPLS-DA) their diagnostic power was found to be limited, as indicated by the area under the curve (AUC) of the receiver operating characteristic (ROC) of 0.77 [17].
In another study, 1H HR-MAS NMR of tissue, in conjunction with 1H NMR from plasma samples, was used to classify papillary thyroid microcarcinomas, a subtype of papillary carcinoma. By using nine significantly changed metabolites from plasma (glucose, mannose, pyruvate, 3-hydroxybutyrate, valine, tyrosine, proline, lysine and leucine), they were able to achieve good sensitivity and specificity with an AUC of 0.992 [18]. This technique was more recently used in FNABs of thyroid tissues collected post-surgically and found statistically relevant metabolites in indeterminate lesions (myo- and scyllo-inositol, serine, citrate, leucine, alanine, phenylalanine and tyrosine) [19]. While HR-MAS NMR can provide 1H NMR spectra of semisolids with a comparable spectral resolution to liquid-state NMR, it requires high spinning rates of several kHz. This may not only disrupt the tissue structure but can also result in the leakage of potentially infectious material. Furthermore, HR-MAS probes are costly, while incomplete suppression of the water signal can also interfere with the quantification of some metabolites. Therefore, 1H-NMR spectroscopy of tissue extracts has continued to be widely used [18][20][21][22][23][24][25]. In the study of Deja et al., four metabolites were considered as selective biomarkers of thyroid cancer, namely creatine, myo- and scyllo-inositol and uracil, but the thyroid cancer group was comprised of only 12 patients [20]. The study by Metere at al., although with only 14 patients, observed differences in cancer and healthy tissue in lactate, phenylalanine, citrate, myo-inositol and threonine [25]. Tian et al. were able to distinguish malignant thyroid lesions from benign with a ROC of 0.88 [21]. On the other hand, Ryoo et al. from the aspirates alone could distinguish seven metabolites (lactate, choline, O-phosphocholine, glycine, citrate, glutamate and glutamine) with ROCs ranging 0.64–0.85 [22]. Seo et al. attempted to predict lymph node metastasis in papillary carcinoma patients, but they were not able to discriminate the presence of metastasis [23]. In the study of Li et al., 15 metabolites were found to be differentiated using two OPLS-DA models [24].
Although NMR spectroscopy has been a valuable technique for several metabolomic studies so far, it has had a strong competition by MS in the last few years. One of the reasons is its lower sensitivity in comparison to MS. However, NMR spectroscopy presents advantages in relation to MS, by being highly reproducible and capable of performing absolute quantification of the metabolite’s concentrations. Furthermore, it can detect compounds that are less easily detected by MS, such as sugars, organic acids, alcohols and other highly polar compounds, and it is well suited for studying intact tissues, organs and other solid or semi-solid samples through solid-state NMR and HR-MAS NMR. However, metabolite identification is not straightforward given the complexity of the 1H-NMR spectra but can be more easily overcome by databases such as the Human Metabolome Database (HMDB) [26], or the use of (semi)automatic identification and quantitation tools such as BAYESIL [27] or Chenomx NMR Suite from Chenomx Inc. This complexity comes mainly from peak overlap, which could be ameliorated by the use of stronger magnets, increasing spectral dispersion. Presently, commercial NMR spectrometers can achieve magnetic fields of 28.2 Tesla, the equivalent to a 1H Larmor frequency of 1.2 GHz, but unfortunately, the cost of such equipment is by now detrimental to their use, with the 600-MHz NMR spectrometers being the best cost-sensitivity/resolution compromise. The more frequent application of selective excitation techniques on specific spectral regions and of multidimensional NMR experiments such as total correlation spectroscopy (TOCSY) and J-resolved spectroscopy (J-Res) [18][21][28] could also help in resolving overlapping peaks. Another exciting development in NMR metabolomics is in probes design, with microprobes for MAS enabling an enhancement of sensitivity while reducing the sample size to a few microliters, and cryoprobes significantly increasing signal sensitivity.
Even though the sensitivity of NMR spectroscopy has significantly improved over the last few years with a metabolite quantification threshold of ≥1 µM, it remains far less than that of MS [29]. With the improvements in instrumentation, experimental methods, software and spectral databases, the use of mass spectrometry in the field of metabolomics has grown considerably in recent years, including its application to metabolomics studies of thyroid cancer (Figure 1). Liquid chromatography coupled to mass spectrometry (LC-MS) was first used to study thyroid cancer in serum samples from 30 papillary thyroid carcinoma, 80 benign thyroid nodules and 30 healthy controls [14]. 3-hydroxybutyric acid, an intermediate product of fatty acid metabolism, was found to be higher in the papillary thyroid carcinoma group compared to either benign or healthy groups. In 2017, Zhou et al. applied a data-independent acquisition (DIA) workflow for metabolomics [30]. Unlike the traditional data-dependent acquisition (DDA) strategies, this acquisition mode has higher metabolite coverage by using mass range windows to obtain the fragmentation spectra. It is expected that this innovative way of LC-MS metabolite profiling will be translated into metabolomic studies [31]. However, alternative approaches can be used in thyroid cancer metabolomics; for example, an amino acid analyser was used on the plasma of thyroid cancer patients and found significantly higher levels of methionine, leucine, tyrosine and lysine [32]. In addition to the analysis of water-soluble metabolites, several lipid species have also been identified as putative biomarkers for resolving malignant and benign thyroid lesions. Ishikawa et al. combined imaging mass spectrometry with a matrix-assisted laser desorption/ionisation tandem time-of-flight (MALDI-TOF/TOF) instrument to identify and describe the distribution of individual biomolecules in a tissue section [33]. With this approach, they revealed that phosphatidylcholine (34:1) and (34:2) and sphingomyelin (34:1) were present in significantly higher amounts in papillary thyroid carcinoma when compared to normal tissue from the same patients. A similar approach was applied to tissue and serum samples collected from subjects with malignant or benign lesions (tissue), as well as healthy subjects with no thyroid lesions (serum). In this case, it was found that a biomarker panel consisting of phosphatidic acid (36:3) and sphingomyelin (34:1) could distinguish malignant cancer from benign, with an AUC value of 0.961, a sensitivity of 87.8% and a specificity of 92.3% [34]. Zhang et al. observed increased relative abundances of ceramides and specific glycerophosphoinositols using 2D desorption electrospray ionisation mass spectrometry to image thyroid cancer in lymph node tissues [35]. Meanwhile, Huanget al. showed a higher expression of phenylalanine, leucine and tyrosine in the tumour region with a gradual level decrease from tumour to the stromal and normal tissues and the inverse profile of creatinine [36]. Another study was able to profile lipids directly in formalin-fixed tissue sections by MALDI-Q-Ion Mobility-TOF-MS, demonstrating that this technique could be complementary to the present histological methods [37]. These studies demonstrate the potential of spatially resolved metabolomics to provide meaningful and clinically relevant information from biopsy samples that are by nature highly heterogeneous.
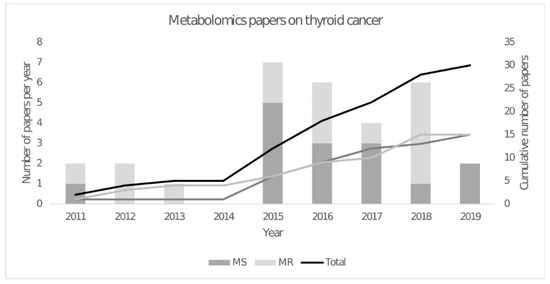
The year 2015 saw a peak in the number of metabolomic publications, with gas chromatography-mass spectrometry (GC-MS) being widely reported (Figure 1). This technique was first used in combination with a 1H NMR metabolomics study to measure fatty acid abundances [21]. They showed higher levels of (C14:0), (C16:0) and (C18:3n3) fatty acids and lower levels of (C20:3n6) fatty acids in malignant compared to benign tissues. Since then, other GC-MS metabolomics studies have been published that identified metabolites in carbohydrate metabolism, including glucose, fructose, galactose, mannose, 2-keto-D-gluconic acid and rhamnose that were decreased in papillary thyroid carcinoma, which is consistent with an upregulation of the glycolysis and pentose phosphate pathways [38]. These results are consistent with cancer tissues requiring higher rates of cytosolic ATP production and increased amounts of NADPH and precursors for biosynthesis of nucleotides and other cell components. Another study combined the metabolic profiles obtained by GC-MS and ultra-performance liquid chromatography-mass spectrometry (UPLC-MS), resulting in a total of 195 detected metabolites. From these metabolites, they concluded that purine and pyrimidine metabolism was higher in papillary thyroid carcinoma, as well as taurine and hypotaurine levels. However, another study that used GC-MS and UPLC-MS identified a decrease in galactinol, melibiose and melatonin in papillary thyroid carcinoma with an AUC of 0.96 [39].
In an attempt to discriminate between different types of thyroid cancer, and some of their most common variants, Wojakowska et al. analysed five different types of thyroid malignancies (follicular, papillary classical variant, papillary follicular variant, medullary and anaplastic cancer), as well as benign follicular adenoma and normal thyroid tissue [40]. They found an upregulation of lactic acid and downregulation of several fatty acids and their esters in cancer versus normal tissue, as well as upregulation of myo-inositol phosphate, succinic acid and certain fatty acids and their esters in malignant versus benign tissue. Moreover, the classical variant of papillary carcinoma could be distinguished from follicular thyroid lesions by lower levels of gluconic acid and higher amounts of citric acid. In addition, follicular carcinoma could be distinguished from the follicular variant of papillary carcinoma by changes in the levels of decanoic acid ester. It would be important to promote more studies which discriminate between different types of thyroid cancer since cancer classification is essential to assess prognosis and select an adequate treatment. Moreover, follicular adenoma, follicular carcinoma and the follicular variant of papillary carcinoma can be hard to distinguish histologically, so metabolomics can represent an important tool to assist in their differentiation. Regarding more specific studies, the serum of 37 patients with distant metastasis was compared with the serum of 40 patients from an ablation group, where it was found that serum asparagine, gamma-amino butyric acid (GABA), aminooxyacetic acid and 4-deoxypyridoxine increased in the distant metastasis group while pyroglutamic acid was decreased [41]. A GC-MS metabolomic study was also performed on a model system of thyrospheres, containing cancer stem-like cells, from B-CPAP and TPC-1 cell lines derived from papillary thyroid cancer of the BRAF-like expression profile class, which showed significant differences in Krebs cycle intermediates, amino acids, cholesterol and fatty acids content when compared to non-cancer stem-like cells [42]. Besides in vivo measurements, it may be interesting to characterise individual cell types found within the tumour, given the heterogeneity of cancer cells, to therapeutically target those that are contributing the most to the cancer phenotype. The papillary thyroid cancer-derived cells also showed altered redox homeostasis as well as increased levels of intracellular oxidant species, a common hallmark of cancer, since ROS homeostasis needs to be tightly regulated, otherwise it can promote an altered metabolism. The most perturbed metabolic phenotype was found in B-CPAP cells, which are characterised by the most aggressive genetic background [43], demonstrating the connection between genetic background and cancer metabolism and consequently phenotype. Once again, we observe the importance of combining information from genetics to metabolism for a better understanding of this disease.
The field of mass spectrometry-based metabolomics has been facing a significant evolution with more sensible, higher dynamic range, higher data acquisition speeds and different acquisition modes equipment. Nonetheless, data acquisition is not the only critical point. Data analysis with better algorithms for peak detection, alignment and analysis; better software tools that integrate these algorithms and further statistical analysis; and better databases with information on each compound, such as possible adducts and multiple retention times (XCMS/Metlin and HMDB), are pushing the field forward at higher speeds. More specifically, identification of metabolites on a large scale with the assistance of software tools (Elucidata El MAVEN [44] and Sciex Accurate Mass Metabolite Spectral Library with MasterView™ software) or using sample preparation kits (IROA® Quantitation Kits) will advance even further mass spectrometry as the go-to methodology for metabolomics. Thyroid cancer profiling, in particular, will definitely benefit from these advances. Moreover, the technical advances for mass spectrometers have allowed even for their use in the clinical setting. Take, for example, an automated and biocompatible handheld mass spectrometer that can quickly and non-destructively assess if at the pointed location cancerous tissue is present, which allows surgeons to accurately define the tumour margins prior to excision [41].
Most of the publications for thyroid cancer metabolomics to date have focused on the direct analysis of the thyroid gland. However, with the aim of avoiding the invasive biopsy of the thyroid, there have also been studies that looked for associations between thyroid malignancies and plasma [12][18][32][45] or serum metabolites [14][34][41][46][47]. A study of children and adolescents with thyroid cancer identified an increase in the levels of serum leucine, lactate, alanine, lysine, acetate, glycine and choline and lower levels of glucose in papillary thyroid carcinoma samples versus benign by 1H NMR spectroscopy, which is consistent with other works in adults [48]. More recently, a plasma GC-MS study has suggested sucrose as a discriminative compound between papillary thyroid cancer and multinodular goitre, which poses an interesting question as to the influence of high sucrose sugar diets in the promotion of tumorigenesis [45]. Another non-invasive approach used capillary electrophoresis to analyse urine [49]. In this case, the authors focused on the profiling of urinary nucleosides, with inosine, N2-methyl guanosine, N2-N2-dimethylguanosine and 1-methylguanosine being higher in thyroid cancer patients when compared to healthy controls. In a study of paired blood and urine samples, it was shown that, while metabolome data of each analyte could differentiate between healthy subjects and those with nodular lesions, analysis of the combined datasets provided better predictive power [50]. Another study, collecting both serum and urine, indicated serum β-hydroxybutyrate, docosahexaenoic acid, 1-methyladenosine, pregnanediol-3-glucuronide, urinary nicotinic acid mononucleotide and xanthosine as a potential biomarker panel for papillary thyroid cancer, using two validation sets [51]. Huang et al. integrated data of serum and plasma metabolites from six independent centres, having had a total of 1540 serum-plasma matched samples and 114 tissues [52]. The study was divided into a discovery phase, composed of one centre and then the validation phase, with the rest of the samples from the other centres. They were able to establish a panel of six biomarkers with an AUC of 98%, namely myo-inositol, α-N-phenylacetyl-L-glutamine, proline betaine, L-glutamic acid, lysophosphatidylcholine (18:0) and lysophosphatidylcholine (18:1), to distinguish between healthy samples and papillary thyroid carcinoma. However, they were not able to distinguish cancer samples from benign thyroid nodules. Another study compared the plasma lipidomic profiles of five commonly found cancers: liver, lung, gastric, colorectal and thyroid [53]. Interestingly, they found a distinct profile in thyroid cancer relative to all the other studied cancers, selecting lysophosphatidylinositol (18:0) and (18:1) as specific to thyroid cancer only. Going beyond blood and urine, a study of thyroid carcinoma patients and healthy controls revealed highly predictive differences in intestinal microbiota genera and faecal metabolites [54]. Finally, exhaled breath from 39 papillary thyroid carcinoma, 25 benign and 32 healthy volunteers was analysed by solid-phase microextraction GC-MS with (3-methyl-oxiran-2-yl)-methanol, 1,1,3-trimethyl-3-(2-methyl-2-propenyl) cyclopentane and trans-2-dodecen-1-ol being identified as significantly changed in papillary thyroid carcinoma versus benign [55].
The study of body fluids is important to give a broader overview of the disease depending on the compartmentalisation of such fluids. For example, urine is highly dependent on food and liquid intake, while blood can have a more stable metabolome. Biofluids imply non-invasive or minimally invasive collection when compared to tissues and reflect the overall response of the patient to the disease. They have therefore the potential to be used in the monitoring of therapy and cancer’s evolution. In the case of thyroid cancer diagnosis, it would be important to find a minor invasive method that could complement the FNAB exam and would ultimately provide a faster and more accurate diagnosis of thyroid cancer. Furthermore, omics profiles in these samples can bring us closer to precision medicine, where an individual’s metabolomic fingerprint can assist the physician in therapy customisation.