Mitostasis refers the mitochondrial dynamics of fusion and fission depending on the cell requirements. Mitostasis also involves mitochondrial traficking and anchoring as need to maintain a functional pool of mitochondria.
1. Introduction
The maintenance and distribution of the mitochondrial reserve is termed mitostasis, which includes mitochondrial transport, anchoring, fission and fusion. In fact, most of the mitochondria do not exist as small, distinct organelles, but rather form a highly interconnected reticulum with contiguous membranes
[1]. This reticulum is highly dynamic with ongoing divisions and reconnections. The balance of fission and fusion determines the length of mitochondria, and it is regulated by stress and nutrient availability
[2]. Mitostasis also involves protein reparation or degradation, and the formation of mitochondria-derived vesicles yielding to mitophagy and macroautophagy
[3].
In particular, mitochondrial biogenesis requires materials from the cytoplasm and nucleus, since mammalian mitochondria DNA (mtDNA) encodes only 13 proteins
[4]. For this reason, mitochondrial biogenesis requires the importation of over 1500 proteins encoded by the nuclear genome
[5] and phospholipids for the inner mitochondrial membrane (IMM) and the OMM
[6].
2. Mitostasis in Neurons
2.1. Mitochondrial Trafficking
The location of mitochondria, depending on local energy demands, is critical for polarized cells such as neurons. Interconnected mitochondria are located in neuronal cell soma. However, to enter the axon, a mitochondrion must undergo a fission reaction. This reaction frees it from its mitochondrial reticulum.
Once released, mitochondrial fast anterograde transport is mediated by microtubules
[7]. Mitochondrial Rho GTPase 1 and 2 (Miro 1 and 2) serve as Ca
2+ sensors that regulate kinesin-mediated mitochondrial motility. Miro 1 and 2 have an outer C-terminal transmembrane domain with GTPase domains, which interact with the trafficking kinesin protein (TRAK)/Milton family of proteins
[8][9][10][11][12]. These cargo adaptorsbind to Kinesin-1 anddynein/dynactin, beingcritical in mitochondrial trafficking and its spatial distribution
[13]. Kinesin-1 cargoes, often together with dynein, drive mitochondria transport into the axon
[11][14], and the dynein/dynactin cargo complextransports mitochondria into dendrites
[13][15].
On the other hand, shorter-range mitochondrial transport is actin-dependent anduses the dynein/dynactin complex as actin-mitochondrial anchorage. The cargo Myosin 19 (Myo19)
[16] has been suggested as critical for this process
[17].
2.2. Mitochondrial Anchoring
Mitochondrial anchoring to inner cell structures is needed to provide the required energy at the right place, as around two-thirds of neuronal mitochondria remain stationary
[18]. These pools of mitochondria are mainly located in the soma and in areas with high ATP demand, such as the nodes of Ranvier, axonal presynaptic endings and dendrites
[19]. Mitochondria anchoring mechanisms are not fully understood and they seem to involve different proteins depending on the localization of the mitochondria. For instance, in axonal presynaptic endings, activation of adenosine monophosphate-activated protein kinase (AMPK) by synaptic activity leads to myosin VI phosphorylation, ending with mitochondrial recruitment and syntaphilin-mediated anchoring on presynaptic filamentous actin
[20]. This process is specific to the stationary mitochondria, not being related to the mitochondria mobility machinery
[21][22]. Remarkably, it is an axon exclusive physiological mechanism. On the other hand, axonal mitochondrial anchor mislocalization to dendrites has been associated with multiple sclerosis disease
[23].
2.3. Mitochondrial Fission
Mitochondrial fission is essential for shaping the mitochondrial network and is required for generating additional mitochondria of the appropriate size for their transport by molecular motors along the cytoskeleton
[24] (
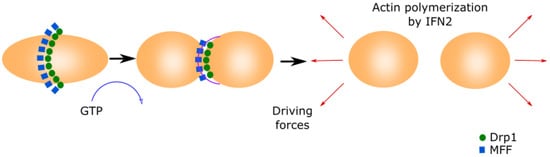
Figure 1).
Figure 1. Mitochondrial fission.This process is initiated by MFF forming a ring in the middle of the primary organelle to be divided. Drp1 GTPase translocates to the mitochondria and binds to MFF, inducing a conformational change by the hydrolysis of GTP. This conformational change generates the ring contraction, which starts mitochondrial fission. The reorganization of the cytoskeleton generates the driving forces that separate both new mitochondria.
Mitochondria fission starts when the mitochondrial fission factor (MFF) surrounds the OMM forming a ring
[25][26]. Mitochondrial fission 1 (Fis1) and mitochondrial dynamics proteins of 49 and 51 kDa (MID49 and MID51) recruit the cytoplasmic Dynamin-related protein 1 (Drp1)
[27][28][29]. Drp1 translocates to the mitochondria and binds to MFF, inducing a conformational change due to its GTPase activity. This conformational change produces the ring contraction that starts mitochondrial fission
[27][30]. Cellular cytoskeleton further separates both mitochondria depending on the MAMs. The Inverted Formin 2 protein (INF2) is localized in MAMs and its activation causes actin polymerization, which generates the driven force separating both mitochondria
[31].
2.4. Mitochondrial Fusion
Mitochondrial fusion is a cell protective mechanism
[32] based in two different and concomitant processes, the OMM fusion and the IMM fusion (
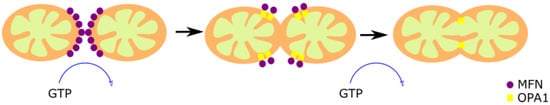
Figure 2).
Figure 2. Mitochondrial fusion. The fusion of two preexistent mitochondria is a protective mechanism based on the fusion of the OMM and IMM. The OMM fusion is mediated by MFN1 and MFN2, and the fusion of the IMM is mediated by OPA1. First, MFN1 and MFN2 GTPase activity induce the formation of homo- or hetero-oligomeric complexes in the fusion spot. Then, OPA1-mediated GTP hydrolysis promotes the membrane fusion.
The dynamin-associated GTPases Mitofusin proteins 1 and 2 (MFN1 and MFN2) are responsible for the OMM fusion
[33][34]. Both GTPases share a high structural and biochemical homology, but they have different roles
[35]. MFN1 is found exclusively in the OMM, and allows the contact between the OMM of different mitochondria
[36]. MFN2, found in mitochondria and ER, has an anchoring role since it joins the two organelles, forming homotypic and heterotypic complexes with MFN1
[37]. The spiral domains of MFN1 and MFN2 interact, forming homo- or hetero-oligomeric complexes through GTP hydrolysis in the fusion spot
[38]. However, there are controversial reports regarding the relevance of this mechanism since it was observed that the loss of MFN-2 increased the ER–mitochondrial juxtaposition
[39]. This fact does not contradict that the functionality of mitochondria–ER contacts is affected in MFN2 KO cells, as suggested by the previous observation that Ca
2+ transfer from the ER to the mitochondria is altered in these cells
[37].
The optic atrophy 1 protein (OPA1) mediates the IMM fusion
[40][41]. The long form of the protein (l-OPA1) works as a coordinator of the IMM of both fusing mitochondria. The two membranes fuse through the short isoform (s-OPA1) by its GTPase activity
[42]. OPA1 is also associated with the maintenance of the MRC by cristae organization, and the control of apoptosis
[43][44][45][46], demonstrating versatility of functions.
2.5. The regulator of Mitochondrial Biogenesis PGC-1α
The peroxisome proliferator-activated receptor-γ (PPARγ) coactivator-1 α (PGC-1α) is a member of a transcription coactivator family, composed ofPGC-1α, PGC-1β and PGC-related coactivator (PRC). These proteins play a central role in the regulation of mitochondrial biogenesis and cellular energy metabolism
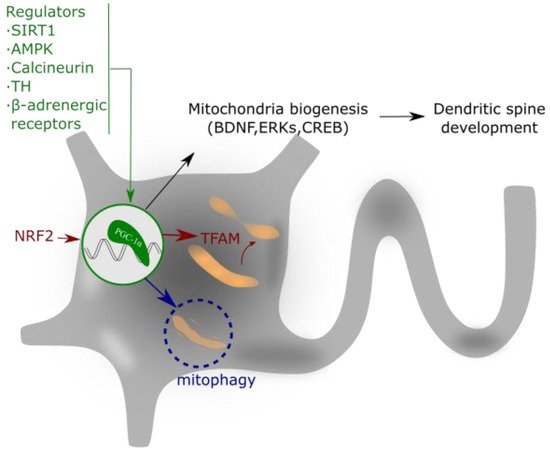
[47] (
Figure 3).
Figure 3. PGC-1α regulates mitochondrial biogenesis and spine growth. Its activity is based on the interaction with NRF that activates to TFAM. This complex regulates mitochondrial gene expression and addresses the requirements of neuronal energy when needed for dendrite activity or growth. PGC-1α expression is regulated by different intracellular signaling pathways, as explained in the text.
The action mechanism of PGC-1α implies its interaction with two nuclear transcription factors, nuclear respiratory factors 1 and 2 (NRF-1 and 2), which activate the mitochondrial transcription factor A (TFAM), a mtDNA-binding protein
[48]. TFAM and the cofactor mitochondrial transcription factor B (TFB1M) are essential for mitochondrial gene expression
[49]. This molecular pathway promotes the equilibrium in the assembly of some components of the MRC, which ultimately ensures appropriate mitochondrial function, mtDNA replication and apoptosis control
[50][51][52].
The expression and/or activity of PGC-1α is regulated by different intracellular signaling pathways such as the PGC-1α regulatory cascade
[53], thyroid hormone (TH)
[54], sirtuin 1 (SIRT1)
[55][56], calcineurin
[57], AMPK
[58], cyclin-dependent kinases (CDKs)
[59], and β-adrenergic signaling
[60]. These regulators of PGC-1α aim to coordinate the expression and function of mitochondrial proteins during mitochondrial biogenesis
[61]. Additionally, not only transcriptional regulation but also post-translational modifications allow the control of the PGC-1α activity. For example, PGC-1α phosphorylation by p38 mitogen-activated protein kinase alpha (p38α) enhances its activity
[62], and PGC-1α acetylation suppresses its activity
[55].
PGC-1α and mitochondrial biogenesis play important roles in the formation and maintenance of dendritic spines and synapses in hippocampal neurons
[63]. In this context, PGC-1α also controls the synthesis of the Brain-Derived Neurotrophic Factor (BDNF)
[64] and the activity of Extracellular Signal-Regulated Kinases (ERKs)
[65]. BNDF has been widely reported in the literature as a neuroprotective agent produced by physical exercise
[66] and, interestingly, exercise induces the upregulation of PGC-1α expression and increases mitochondrial mass within the brain
[67]. In addition to its role in mitochondrial biogenesis, PGC-1α also regulates apoptosis and the ROS detoxification system
[68].
3. Conclusions
Aging and neurodegenerative processes are not the consequences of a unique signaling pathway alteration but of multifactorial events, including dysfunction of mitochondria that yield to failures in ATP production, increased ROS production and alterations in Ca2+ homeostasis. Considering mitochondria as a therapeutic target would produce an improvement in the maintenance of the optimum neuronal function. These therapeutic approaches would include the regulation of PGC-1α and calcium buffering systems as well as free radicals’generation, addressing the source of free radicals more than the use of antioxidants that have not demonstrated effective results to date.