1000/1000
Hot
Most Recent

Triple Negative breast cancer is unresponsive to endocrine therapy, i.e., tamoxifen, aromatase inhibitors, and/or anti-HER2-targeted therapies. As there are currently no other therapeutic options to treat TNBC apart from chemotherapy, various studies were reviewed to draw the conclusion that ncRNAs might be candidates for drug development and drug resistance. Targeted approaches to epigenetic mechanisms and clarification of the molecular mechanisms of specific miRNAs in TNBC subtypes are fully justified. Currently, TNBC with BLP is the only subtype for which we might suggest some therapeutic hypotheses based on miRNA/mRNA integration. This review might provide a collection of biomarkers potentially useful in clinical settings and shows that the combination of miRNA-based therapeutic strategies with conventional therapies might synergize anticancer effects improving patient outcome.
Ten to twenty percent of invasive breast cancer (BC) belong to the TNBC type, which is prevalent in young women <50 years of age; among African, American, and Hispanic women; and in women with higher premenopausal body mass index, earlier age at menarche, and higher parity. Specifically, the Triple Negative phenotype (TNP) is defined by ER/PR/HER2-negative immunostaining, habitually with higher expression of the Ki-67 antigen, higher mitotic index, and BRCA1 gene mutations (about 75%). TNBC shows aggressive behavior with high histological grade, high recurrence rate in about 2 or 3 years after treatment compared with other BC subtypes, and frequent distant metastasize; thus, it accounts for about 25% of BC-related deaths[1][2][3][4][5]. TNBC shows different molecular and clinicopathological features[6] and is histologically categorized as a high-grade invasive BC of no special type. “Special types” are still included into the TNBC subtype but differ in biological behavior and clinical course[7].
BLBC is characterized by gene expression usually found in basal or myoepithelial mammary cells, shown by high molecular weight cytokeratin, prevalently CK5/6, and by EGFR expression, with about 75% of them referred to as TNP, with ER/PR/HER2 also being negative by immunohistochemistry (IHC). The remaining 25% includes all other “intrinsic” BC molecular subtypes. BLBC shows short survival following progression to metastatic disease, with prevalence of cerebral and lung metastases with respect to luminal subtypes[8][9]. Remarkably, 80% of TNBCs show basal-like features: TNBC and BLBC phenotypes are effectively synonymous [10], although immunohistochemical expression, transcriptomic, and clinical data suggest they are not equivalent[11][12].
The lack of hormonal and HER2 receptors makes TNBC unresponsive to endocrine therapy, i.e., tamoxifen, aromatase inhibitors, and/or anti-HER2-targeted therapies, so surgery, radiotherapy, and mostly nonspecific chemotherapies (e.g., anthracycline and taxane regimens) remain the mainstay for management of these patients, often with severe side effects affecting life quality [5][13]. TNBC treatment is a major challenge for oncologists, both due to heterogeneity of disease and to the absence of unambiguous molecular targets.
MicroRNAs (miRNAs) are a family of endogenous, short single-stranded, noncoding RNA regulating gene expression by interacting with complementary mRNA-target sequences causing either mRNA degradation or translational repression[14]. MiRNAs regulate multiple processes[14][15], and their dysregulation is strongly related to cancer involving alteration of biological functions [16]. MiRNA level variations were analyzed comparing normal vs. neoplastic tissues[17][18] in several BC subtypes[19]with different responses to endocrine therapy [20]. Several miRNAs were related to pathogenesis of TNBC[21][22] and might represent potential predictors of anticancer drugs efficacy and prognosis[23][24][25]. Recognizing gene expression regulator-classified miRNAs as epigenetic elements involved in cancer development means that they might be potentially used as therapeutic targets and diagnostic/prognostic biomarkers in order to achieve high accuracy in tumor classification[26].
TNBC morphological and molecular heterogeneity, poor prognosis, and lack of specific targeted therapies require a detailed understanding of biology and classification in this type of cancer. Gene expression profiling based on high-throughput technologies provided the basis for an improved TNBC molecular taxonomy, establishing prognostic and predictive indicators. The adoption of unsupervised clustering analysis led to identification of several TNBC molecular subtypes, showing transcriptomic heterogeneity and unique biological pathways.
Lehmann et al. showed that TNBCs include different molecular subtypes, i.e., basal-like 1-2 (BL1-2), immunomodulatory (IM), claudin-low-enriched mesenchymal (M), mesenchymal stem-like (MSL), and luminal androgen receptor (LAR), each one showing a distinctive biology and drug sensitivities[27]. Gene ontology (GO) for the BL-1 subtype is enriched in cell-cycle, cell-division, and DNA damage response (ATR/BRCA) pathways, confirming increased proliferation and loss of cell-cycle checkpoints. BL-2 GO is improved in growth factor receptors (EGFR, MET, and EPHA2) showing specific characteristics of basal/myoepithelial origin with high expression of TP63 and MME mRNAs as well as in glycolysis and gluconeogenesis. The IM subtype is enhanced in immune cell processes (cell and cytokine signaling, antigen processing, and presentation) and signaling by transduction of the immune signal of the nucleus. IM GO overlaps with a gene signature for medullary BC, an unusual, high-grade histologic TNBC subtype showing favorable prognosis[7][28]. The M and MSL subtype GOs are enriched in cell motility, extracellular matrix (ECM) receptor interaction, and cell differentiation. Additionally, the MSL subtype is involved in cell growth and angiogenesis while expresses low levels of proliferation genes with expression enrichment in stem cells, HOX genes, and mesenchymal stem cell-specific markers. The pathway components in the M and MSL subtypes accounts for highly dedifferentiated metaplastic BC, featuring mesenchymal/sarcomatoid or squamous characteristics and chemoresistance[28]. The MSL subtype showed low levels of claudins 3, 4, and 7, congruent with the BC claudin-low subtype [29]. Claudin-low-expressing tumors exhibit a high expression of genes associated with epithelial–mesenchymal transition (EMT). The LAR subtype, although being ER negative, shows a luminal-like expression profile, with a strong expression of AR and downstream AR targets and coactivators. GO is enriched in hormonal-regulated pathways inclusive of steroid synthesis, porphyrin metabolism, and androgen/estrogen metabolism. The LAR subtype consists of AR-driven tumors that encompass the molecular apocrine type.
BC classification as a luminal or basal-like subtype is based on typical protein expression. Luminal BC subtypes express the protein of luminal epithelial cells, called the ‘‘luminal group”, such as luminal cytokeratins (CK8 and 18), ER and GATA3; BLBCs express high molecular weight basal cytokeratins (CK5/6, 14, and 17) and EGFR and/or c-KIT[30]. In TNBC, the BL1-2 and M subtypes express high levels of basal cytokeratin, while the LAR subtype expresses high levels of luminal cytokeratins and luminal markers (FOXA1 and XBP1). To complete the previous BC classification, the intrinsic molecular subtype of BC[31] compared to TNBC subtypes[27] established that 49% of TNBC can be classified as basal-like. A strong association was identified between the BLBC subtype and BL-1 TNBC subtypes, while the BL2, IM, and M subtypes were moderately related to the basal-like molecular class [27]. Finally, a total of 82% of LAR subtypes was classified as Luminal A or B and none were classified as basal-like, strengthening the luminal origin.
Prat et al. showed higher concordance between the TNBC and BLBC subtypes on which was applied the PAM50 intrinsic subtype classifier, finding 78.6% BLP, 7.8% HER2-enriched, 6.6% luminal, and 7% normal-like phenotypes in accordance with three wide clinical trials reviewed by IHC-based and PAM50-based data. Contrariwise, in BLBC, 68.5% was ER−/HER2−, 18.2% was ER+/HER2−, 10.6% was ER−/HER2+, and 2.7% was ER+/HER2+[12]. The expression profiles in the luminal, HER2-enriched, and basal-like subtypes revealed six main gene clusters[12]. Unsurprisingly, Triple Negative (TN)/luminal exhibited high levels of estrogen-related and luminal genes and low levels of cell cycle-related genes. TN/basal-like included basal epithelial cell and proliferation genes. TN/HER2+ showed increased expression of genes related to oxidation reduction-related biological activities. Moreover, a subcluster of luminal-like genes involving AR was also identified in TN/luminal and TN/HER2+. Although TNBC is biologically heterogeneous, the expression profile of intrinsic molecular subtypes is preserved and no strong differences were observed between intrinsic molecular subtypes with and without TNP[12].
Using the fuzzy clustering method, an unsupervised analysis of gene expression profiles identified three TNBC clusters: C1 (22.4%), C2 (44.9%), and C3 (32.7%). C1 represented a no basal-like phenotype, enriched in luminal and AR genes. C2 was considered pure basal-like. C3 was enriched in BLP including 26% of claudin-low subtypes, and M2-like macrophages were a hallmark of C3[32].
Burstein et al. defined four TNBC subtypes with unique copy number variations (CNVs) and distinct clinical outcomes. The LAR subtype exhibits the involvement of ER and the PR, FOXA, XBP1, and GATA3 genes, suggesting the evidence of ER activation in “ER-negative” tumors that might be related to 1% of cancer cells showing low levels of ER protein. LAR tumors may respond to traditional anti-estrogen therapies as well as to anti-androgens. The Mesenchymal (MES) subtype is characterized by deregulation of cell cycle, mismatch repair, DNA damage, and hereditary BC signaling pathways, such as overexpression of genes restricted to osteocytes (OGN) and adipocytes (ADIPOQ and PLIN1) and essential growth factors (IGF-1). Basal-Like Immune-Suppressed (BLIS) reveals downregulation of B, T, and NK cell immune-regulating and cytokine networks, showing the worst prognosis. Conversely, Basal-Like Immune-Activated (BLIA) exhibits upregulation of the genetic factor controlling B, T, and NK cell functions; activation; and high expression of STAT pathways and shows the best prognosis[33].
Le Du et al. revised TNBC molecular classifications and defined five potential clinical groups. BLBC is the most frequent subtype (25–80% of cases), characterized by DNA-repair gene deficiency and/or growth factor pathway overexpression, showing the highest pathological response (pCR) to chemotherapeutic treatment and sharing this feature with the Lehman-BL-1 group. They include the Lehman-BL-2 group into the mesenchymal subtype because it is enriched in growth factor/receptor tyrosine kinase pathways. The mesenchymal-like subtype includes the mesenchymal, mesenchymal stem-like, and claudin low subtypes and is characterized by EMT and cancer stem cell (CSC) characters, correlated with chemotherapy resistance[34]. The immune response signature was correlated with enriched levels of immune cell infiltration and good clinical response[35]. Tumor-infiltrating lymphocytes predict neoadjuvant response to chemotherapy[36][37]. Luminal/apocrine is enriched in hormonal-regulated pathways: AR overexpression might supply a lack of ER expression in steroid-signaling[37]. This group could include the LAR, Luminal A-B, Burstein’s LAR, and molecular apocrine subtypes, showing high expression of luminal gene, lack of basal-cytokeratin markers, and low proliferation rate[12][27]. The AR positivity, identified by IHC in a minimum of 10% of cancer cells, is predictive of about 30% of TNBCs with favorable prognosis[38][39][40] Finally, they proposed the HER2-enriched subgroup as a distinct clinical entity but hypothesized the possibility that Luminal A and HER2-enriched subtypes can be brought together in a short period of time and suggested evaluating HER2-targeted therapies for this group of patients.
In 2016, the TNBC subtypes were refined by using histopathological quantification, laser capture microdissection, and RNASeq. The IM and MSL subtypes were excluded as TNBC subtypes as their features were associated to tumor-associated stromal cells and infiltrating lymphocytes and not to tumor cells[41]. Different clinical features and progression patterns have been demonstrated among the four TNBC subtypes. BL1 displays higher grade, lower stage, and amplified relapse-free overall survival (OS) of patient. LAR shows wider regional diffusion and preferentially distant metastasis to bone, whereas M tumors show that to lung. TNBC subtypes diverge in response to conventional neoadjuvant chemotherapy. The BL1 subtype displays the highest likelihood of getting a pCR, while LAR subtypes showed improved outcome against a reduced answer to neoadjuvant chemotherapy that could be caused by reduction in proliferation and luminal state. The clinical utility to stratify TNBC patients is that it could lead to selection of patients more responsive to chemotherapy [41].
Prado-Vazquez et al. analyzed clinical and expression profile data of TNBC through hierarchical clustering and probabilistic graphical models (PGM) showing two TNBC molecular classifications based on cellular type and immune activity. Based on the PGM, they distinguished four subgroups: CLDN-low, CLDN-high, basal-like, and LAR, consistent with the CSC hypothesis[42]. These classes define the differentiation process where the stem cell becomes carcinogenic and underline that CLDN-low is the less differentiated cancer, with respect to LAR, which is the most differentiated one. Strong molecular differences for therapeutic utility were identified. CLDN-low showed low alpha-amylase activity and regulation of actin cytoskeleton, and high haptoglobin activity. CLDN-high had low actin binding and high chemokine activity. Basal tumors had high cell adhesion and regulation of the actin cytoskeleton activity. LAR tumors showed low intensity regarding cell adhesion, G1/S transition of mitotic cell cycle, and chemokine action. Two additional TNBC subgroups have been defined, immune-positive and immune-negative, and demonstrated that the immune activation was significantly associated with good prognosis and positively influences the prognosis in the cellular LAR and CLDN-high groups[42].
Figure 1 illustrates the high molecular heterogeneity of TNBC as described by the cited authors. It shows the close relationship between TNBC and the basal-like phenotype, which is attested in each TNBC classification study, and the possibility to identify TNBC with other intrinsic phenotypes.
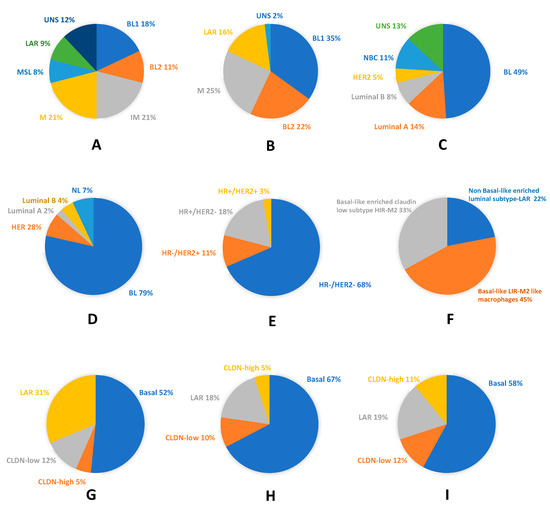
Figure 1. Molecular and phenotypic heterogeneity of Triple Negative breast cancer (TNBC): (A) pie chart showing frequencies and molecular TNBC type-6 classification (according to Lehmann et al. 2011)[13]; (B) pie chart displaying frequencies and molecular TNBC type-4 classification (according to Lehmann et al. 2011)[41]; (C) TNBC assigned to intrinsic molecular breast cancer (BC) subtypes (according to Lehmann et al. 2011)[13]; (D) pie chart showing molecular subclassification and frequencies of basal-like phenotype (BLP) in TNBC (according to Prat et al. 2013)[44]; (E) pie chart displaying molecular subclassification and frequencies of TNP in BLBC (according to Prat et al. 2013) [44]; (F) pie chart showing the fuzzy clustering of TNBC (according to Jezequel et al.) [45]; (G) pie chart displaying the probabilistic graphical model of TNBC (according to Prado-Vazquez et al.)[42]; (H) pie chart showing the overlapping between the probabilistic graphical model of TNBC and the immune activity negative group (according to Prado-Vazquez et al.)[42]; and (I) pie chart displaying the overlapping between the probabilistic graphical model of TNBC and the immune activity-positive group (according to Prado-Vazquez et al.)[42]. BL, basal-like; BL1, basal-like 1; BL2, basal-like 2; CLDN, Claudin; HER2, human epidermal growth factor receptor 2; HR−, hormone receptor negative; HR+, hormone receptor positive; IM, immunomodulatory; LAR, luminal AR; M, mesenchymal; MSL, mesenchymal stem-like; NBC, non-basal-like; NL, normal-like; and UNS, unstable.
Molecular differences identified in TNBCs might be valuable for biomarker identification, drug discovery, and clinical trial design and could support the variations in histological types, prognosis, and patient’s outcome characterizing each TNBC molecular subtype. Notwithstanding some divergences, the TNBC molecular classifications previously discussed provide adequate evidence that there are four major subtypes demanding subtype-specific biological-based therapies. Predicted “driver” signaling pathways were pharmacologically targeted in clinical trials to prove if gene expression signatures can impact therapy selection (Figure 2).
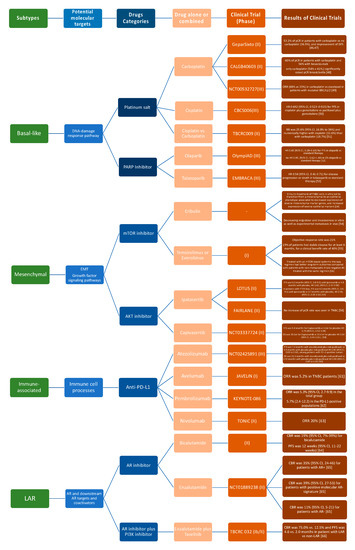
Figure 2. Current advances in systematic treatment for patients affected by different TNBC subtypes [46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66].
Several studies have been performed to analyze clinical differences among TNBC molecular subtypes proving that BLP is of particular clinical interest, being diagnosed in younger women and showing higher grade, advanced clinical disease, and higher stage relative to non-basal TNBC. Although there are no differences in regional spread to lymph nodes and brain/lung metastasis between basal and non-basal TNBC[41], nonetheless, the M subtype displays a significantly higher frequency of lung metastasis vs. all other subtypes; significant incidence of bone metastasis has been identified in the LAR subtype, in accordance with the preference of hormonal-regulated cancers to metastasize to bone[67]. Basal-like TNBCs are largely classical ductal carcinomas, in contrast with lobular carcinomas, which are nearly exclusive to the LAR subtype, suggesting a potential role for AR signaling in lobular BC. Medullary histological types are common in all TNBC subtypes except for the M subtype, consistent with the lack of lymphocytic infiltration in these tumors. Metaplastic carcinomas are typical of basal-like and M subtypes. Specifically, the IM subtype displays the highest number of lymphocytes and lower involvement of lymph nodes, showing consequently the best OS and release-free survival (RFS). The LAR subtype is diagnosed in women of older ages compared to all other TNBC subtypes, showing lower grade and significant enrichment of lymph node metastasis[41]. Rakha et al. showed shorten BC-specific survival and shorter disease-free survival (DFS) in patients affected by TNBC with basal-markers matched to those with TNBC without basal-markers[68].
Strategies to reprogram aberrant miRNA networks in cancer would be more effective considering that proliferation, invasion-migration, and acquisition of CSC properties represent biological processes to improve cancer progression and poor clinical patient’s outcome. Between the TNBC molecular classes, more genomic-epigenomic variations have been identified for the basal-like subtypes; Figure 3 represents a miRNAs/mRNA complex network between pathways involved in growth; proliferation; apoptosis; and processes such as EMT, CSC properties, and suppression of ER mRNA levels involved in the development of this aggressive BC subtype. While mRNA drugs inhibitors are well-known, as described in the figure, very little is known about its mechanisms and drug development to control oncomiR/anti-oncomiR. Currently, therapeutic methods to silence/activate oncomiR/anti-oncomiR have been hypothesized and studied, such as epigenetic silencing of cognate host genes, development of antagomirs or mimic-miRNAs conveyed by nanoparticles, and identification of specific transcription factors for miRNAs which could be hypothesized to modulate miRNA expression variations in cancer.
Figure 3. miRNAs/mRNA complex network in TNBC with basal-like phenotype and potential sites of therapeutic interventions.