Nanobiotechnology involves the study of structures found in nature to construct nanodevices for biological and medical applications with the ultimate goal of commercialization. Within a cell most biochemical processes are driven by proteins and associated macromolecular complexes. Evolution has optimized these protein-based nanosystems within living organisms over millions of years. Among these are flagellin and pilin-based systems from bacteria, viral-based capsids, and eukaryotic microtubules and amyloids. While carbon nanotubes (CNTs), and protein/peptide-CNT composites, remain one of the most researched nanosystems due to their electrical and mechanical properties, there are many concerns regarding CNT toxicity and biodegradability. Therefore, proteins have emerged as useful biotemplates for nanomaterials due to their assembly under physiologically relevant conditions and ease of manipulation via protein engineering.
1. Introduction
The term bionanotechnology refers to the use of biological molecules engineered to form nanoscale building materials. The assembly of small molecules into more complex higher ordered structures is referred to as the “bottom-up” process, in contrast to nanotechnology which typically uses the “top-down” approach of producing smaller macroscale devices. These biological molecules include DNA, lipids, peptides, and more recently, proteins. The intrinsic ability of nucleic acid bases to bind to one another due to their complementary sequence allows for the creation of useful materials. It is no surprise that they were one of the first biological molecules to be implemented for nanotechnology
[1][2][3][4]. Similarly, the unique amphiphilicity of lipids and their diversity of head and tail chemistries provide a powerful outlet for nanotechnology
[5]. Peptides are also emerging as intriguing and versatile drug delivery systems (recently reviewed in
[6]), with secondary and tertiary structure induced upon self-assembly. This rapidly evolving field is now beginning to explore how whole proteins can be utilized as nanoscale drug delivery systems
[7]. The organized quaternary assembly of proteins as nanofibers and nanotubes is being studied as biological scaffolds for numerous applications. These applications include tissue engineering, chromophore and drug delivery, wires for bio-inspired nano/microelectronics, and the development of biosensors.
The molecular self-assembly observed in protein-based systems is mediated by non-covalent interactions such as hydrogen bonds, electrostatic, hydrophobic and van der Waals interactions. When taken on a singular level these bonds are relatively weak, however combined as a whole they are responsible for the diversity and stability observed in many biological systems. Proteins are amphipathic macromolecules containing both non-polar (hydrophobic) and polar (hydrophilic) amino acids which govern protein folding. The hydrophilic regions are exposed to the solvent and the hydrophobic regions are oriented within the interior forming a semi-enclosed environment. The 20 naturally occurring amino acids used as building blocks for the production of proteins have unique chemical characteristics allowing for complex interactions such as macromolecular recognition and the specific catalytic activity of enzymes. These properties make proteins particularly attractive for the development of biosensors, as they are able to detect disease-associated analytes in vivo and carry out the desired response. Furthermore, the use of protein nanotubes (PNTs) for biomedical applications is of particular interest due to their well-defined structures, assembly under physiologically relevant conditions, and manipulation through protein engineering approaches
[8]; such properties of proteins are difficult to achieve with carbon or inorganically derived nanotubes. For these reasons, groups are studying the immobilization of peptides and proteins onto carbon nanotubes (CNTs) in order to enhance several properties of biocatalysis such as thermal stability, pH, operating conditions etc. of the immobilized proteins/enzymes for applications in bionanotechnology and bionanomedicine. The effectiveness of immobilization is dependent on the targeted outcome, whether it is toward high sensitivity, selectivity or short response time and reproducibility
[9]. A classic example of this is the glucose biosensor
[10][11], where glucose oxidase (GOx) is immobilized onto CNTs, for detection of blood glucose levels; this approach can also be adapted for the development of GOx-CNT based biocatalysis for micro/nanofuel cells for wearable/implantable devices
[9][12][13][14].
The use of proteins for the de novo production of nanotubes continues to prove quite challenging given the increased complexity that comes with fully folded tertiary structures. As a result, many groups have looked to systems found in nature as a starting point for the development of biological nanostructures. Two of these systems are found in bacteria, which produce fiber-like protein polymers allowing for the formation of extended flagella and pili. These naturally occurring structures consist of repeating monomers forming helical filaments extending from the bacterial cell wall with roles in intra and inter-cellular signaling, energy production, growth, and motility
[15]. Another natural system of interest has been the adaptation of viral coat proteins for the production of nanowires and targeted drug delivery. The artificial modification of multimer ring proteins such as wild-type
trp tRNA-binding attenuating protein (TRAP)
[16][17][18],
P. aeruginosa Hcp1
[19], stable protein 1 (SP1)
[20], and the propanediol-utilization microcompartment shell protein PduA
[21], have successfully produced nanotubes with modified dimensions and desired chemical properties. We discuss recent advances made in using protein nanofibers and self-assembling PNTs for a variety of applications.
2. Protein Nanofibers and Nanotubes (NTs) from Bacterial Systems
Progress in our understanding of both protein structure and function making up natural nanosystems allows us to take advantage of their potential in the fields of bionanotechnology and nanomedicine. Understanding how these systems self-assemble, how they can be modified through protein engineering, and exploring ways to produce nanotubes in vitro is of critical importance for the development of novel synthetic materials.
2.1. Flagella-Based Protein Nanofibers and Nanotubes
Flagella are hair-like structures produced by bacteria made up of three general components: a membrane bound protein gradient-driven pump, a joint hook structure, and a long helical fiber. The repeating unit of the long helical fiber is the FliC (flagellin) protein and is employed primarily for cellular motility. These fibers usually vary in length between 10–15 µm with an outer diameter of 12–25 nm and an inner diameter of 2–3 nm. Flagellin is a globular protein composed of four distinct domains: D0, D1, D2, and D3
[22]. The D0, D1 and part of the D2 domain are required for self-assembly into fibers and are largely conserved, while regions of the D2 domain and the entire D3 domain are highly variable
[23][24], making them available for point mutations or insertion of loop peptides. The ability to display well-defined functional groups on the surface of the flagellin protein makes it an attractive model for the generation of ordered nanotubes. Up to 30,000 monomers of the FliC protein self-assemble to form a single flagellar filament
[25], but despite their length, they form extremely stiff structures with an elastic modulus estimated to be over 10
10 Nm
−2 [26]. In addition, these filaments remain stable at temperatures up to 60 °C and under relatively acidic or basic conditions
[27][28]. It is this durability that makes flagella-based nanofibers of particular interest for applications that require harsh environmental conditions.
Initial adaptation of the flagellar system for bionano applications targeted
E. coli flagellin, where thioredoxin (
trxA) was internally fused into the
fliC gene, resulting in the FliTrx fusion protein
[29]. This fusion resulted in a partial substitution of the flagellin D2 and D3 domains, with TrxA being bounded by G243 and A352 of FliC, importantly keeping the TrxA active site solvent accessible. The exposed TrxA active site was then used to introduce genetically encoded peptides, including a designed polycysteine loop, to the FliTrx construct. Since the domains responsible for self-assembly remained unmodified, flagellin nanotubes formed having 11 flagellin subunits per helical turn with each unit having the ability to form up to six disulfide bonds with neighboring flagella in oxidative conditions. Flagella bundles formed from these Cys-loop variants are 4−10 µm in length as observed by fluorescence microscopy and represent a novel nanomaterial. These bundles can be used as a cross-linking building block to be combined with other FliTrx variants with specific molecular recognition capabilities
[29]. Other surface modifications of the FliTrx protein are possible by the insertion of amino acids with preferred functional groups into the thioredoxin active site. Follow-up studies by the same group revealed a layer-by-layer assembly of streptavidin-FliTrx with introduced arginine-lysine loops producing a more uniform assembly on gold-coated mica surfaces
[30].
Flagellin is increasingly being explored as a biological scaffold for the generation of metal nanowires. Kumara et al.
[31] engineered the FliTrx flagella with constrained peptide loops containing imidazole groups (histidine), cationic amine and guanido groups (arginine and lysine), and anionic carboxylic acid groups (glutamic and aspartic acid). It was found that introduction of these peptide loops in the D3 domain yields an extremely uniform and evenly spaced array of binding sites for metal ions. Various metal ions were bound to suitable peptide loops followed by controlled reduction. These nanowires have the potential to be used in nanoelectronics, biosensors and as catalysts
[31]. More recently, unmodified
S. typhimurium flagella was used as a bio-template for the production of silica-mineralized nanotubes. The process reported by Jo and colleagues in 2012
[32] involves the pre-treatment of flagella with aminopropyltriethoxysilane (APTES) absorbed through hydrogen bonding and electrostatic interaction between the amino group of APTES and the functional groups of the amino acids on the outer surface. This step is followed by hydrolysis and condensation of tetraethoxysilane (TEOS) producing nucleating sites for silica growth. By simply modifying reaction times and conditions, the researchers were able to control the thickness of silica around the flagella
[32]. These silica nanotubes were then modified by coating metal or metal oxide nanoparticles (gold, palladium and iron oxide) on their outer surface (
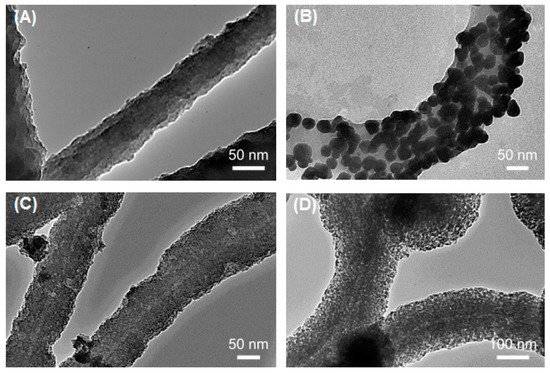
Figure 1). It was observed that the electrical conductivity of the flagella-templated nanotubes improved
[33], and these structures are currently being investigated for use in high-performance micro/nanoelectronics.
Figure 1. Transmission electron microscope (TEM) micrographs of pristine and metalized Flagella-templated silica nanotubes. (
A) Pristine silica nanotubes fabricated on flagella bio-templates. (
B) Gold, (
C) palladium, and (
D) iron oxide nanoparticles deposited on the silica nanotubes. (Reprinted with permission from Jo et al.
Nanotechnology 24, 13574 (2013)
[33]).
2.2. Pilin-Based Protein Nanotubes
Type 4 Pili (T4P) are polymers of a single monomeric type IV pilin subunit that extends from the surface of gram-negative bacteria to form fiber-like structures with a length ranging several micrometers and a diameter of approximately 6 nm
[34][35][36]. Bacteria utilize T4P to mediate a variety of biological processes including cell-host attachment, microcolonization, biofilm formation, and twitching motility
[37][38][39][40][41]. Atomic models for pilins from several bacteria have been characterized including, among others, pilins from
P. aeruginosa strains PAK
[42][43], K122-4
[44][45], PAO
[46], Pa110594
[47],
Neisseria gonorrhoeae strain MS11
[48],
Clostridium difficile [49][50], and the toxin coregulated pilin (TcpA) of
Vibrio cholerae [36]. Pilin proteins are comprised of a long N-terminal α-helix, a four-stranded antiparallel β-sheet with connecting loops, and a C-terminal disulfide bounded receptor-binding D-region
[15]. The assembly of T4P has been well studied; all T4P models place the hydrophobic N-terminal α-helix in the interior of the pilus while the variable β-sheets are exposed on the outer surface
[51]. Thus, the N-terminal α-helix is protected from the immune system and acts as a conserved oligomerization domain
[8][15][45]. Recent work on the K122-4 pilin from
P. aeruginosa has revealed that the protein oligomerizes into nanotubes in the presence of hydrophobic surfaces or compounds (
Figure 2)
[52][53][54][55]. While generated in vitro, the pilin-derived PNTs share a similar morphology and diameter (~5–6 nm) to in vivo T4P
[52][53][54], the former can reach a length of several hundred micrometers compared to native pili that typically have a length of 10 µm
[34][35][36][51].
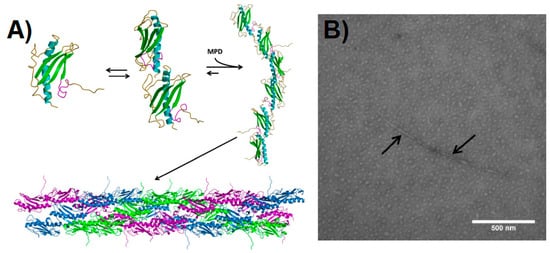
Figure 2. Pilin-derived protein nanotube (PNT) assembly. (
A) The ΔK122 pilin (PBD ID 1QVE
[45]) exists as a monomer-dimer equilibrium in solution
[55]. The common structural features of the type IV pilins are highlighted in the monomer—the N-terminal α-helix in cyan, the β-sheet in green, coil regions in gold, and the receptor-binding domain (known to mediate surface interactions) in magenta. Upon addition of a hydrophobic compound such as 2-methyl-2,4-pentanediol (MPD), the ΔK122 pilin forms fibrils that can then assemble into PNTs. The three ΔK122 fibrils observed in a helical assembly of native T4P are shown in purple, green, and blue, respectively. (
B) Upon the addition of the oligomerization initiator MPD, the ΔK122 monomer/dimers are seen as aggregates in TEM, and form pilin fibrils (highlighted by arrows). (Reprinted with permission from Petrov et al.
J. Nanobiotechnol.
11, 24 (2013)
[54]).
From a bionanotechnology perspective, T4P form robust nanofibers with the ability to bind biotic and abiotic surfaces via their tips. These interactions have been mapped to the D-region of the pilin. It has been estimated that the attractive force between the native T4P tip and steel is in the range of 26–55 pN/molecular interaction and for in vitro derived nanotubes is in the range of 78–165 pN/molecular interaction
[56]. Functional nanostructures have been generated from native bacterial pili and explored for their potential use as biological nanowires. For example, the type IV pili of
Geobacter sulfurreducens reduces Fe(III) oxides by transporting electrons over long distances and has potential applications for use in microbial-based fuel cells
[57][58]. Further studies have shown that cultures of
G. sulfurreducens produce biofilms that exhibit high current densities—one of the highest known current densities when incorporated into microbial fuel cells
[59]. These
G. sulfurreducens pili are capable of long-range metallic-like conductivity
[60] and supercapacitor behavior
[61], making them an exciting prospect for use as a low-cost and environmentally sustainable form of energy storage.
The β-sheet and connecting loops of the type IV pilins form the surface of the pilus, and are thus exposed to the immune system. As a result these regions show significant sequence variability between bacterial systems. This allows for the use of mutagenesis to design fibers with altered surface properties. Research is ongoing to explore how protein engineering of the monomer can lead to nanofiber attachment to other abiotic surfaces. For instance, addition of a polyhistidine tag to the C-terminus of the protein can potentially direct binding to nickel and copper surfaces or nanoparticles. If we consider binding of T4P/PNT to biotic surfaces such as epithelial cells, this opens an exciting area for further research in therapeutics. As is the case with binding to abiotic surfaces, the D-region of the pilin is responsible for forming specific interactions with cellular glycolipids
[62]. This receptor-specific interaction can allow for mediated drug delivery upon binding of the synthetic nanofibers.
3. Virus-Based Protein Nanotubes (PNTs)
Viral capsids are protein shells that serve to protect the enclosed genetic material. These self-assembling capsids are formed from relatively simple protein building blocks making them ideal for the production of nanostructures. Capsids vary in size from 18–500 nm with morphologies ranging from helical (rod-shaped) to icosahedral (spherical-shaped). These structures can be chemically and genetically manipulated to fit the needs of various applications in biomedicine, including cell imaging and vaccine production, along with the development of light-harvesting systems and photovoltaic devices. Due to their low toxicity for human applications, bacteriophage and plant viruses have been the main subjects of research
[63]. Below, we highlight three widely studied viruses in the field of bionanotechnology.
3.1. Tobacco Mosaic Virus (TMV)
The concept of using virus-based self-assembled structures for use in nanotechnology was perhaps first explored when Fraenkel-Conrat and Williams demonstrated that tobacco mosaic virus (TMV) could be reconstituted in vitro from its isolated protein and nucleic acid components
[64]. TMV is a simple rod-shaped virus made up of identical monomer coat proteins that assemble around a single stranded RNA genome. RNA is bound between the grooves of each successive turn of the helix leaving a central cavity measuring 4 nm in diameter, with the virion having a diameter of 18 nm. It is an exceptionally stable plant virus that offers great promise for its application in nanosystems. Its remarkable stability allows the TMV capsid to withstand a broad range of environments with varying pH (pH 3.5–9) and temperatures up to 90 °C for several hours without affecting its overall structure
[65]. Early work on this system revealed that polymerization of the TMV coat protein is a concentration-dependent endothermic reaction and depolymerizes at low concentrations or decreased temperatures. According to a recent study, heating the virus to 94 °C results in the formation of spherical nanoparticles with varying diameters, depending on protein concentration
[66]. Use of TMV as biotemplates for the production of nanowires has also been explored through sensitization with Pd(II) followed by electroless deposition of either copper, zinc, nickel or cobalt within the 4 nm central channel of the particles
[67][68]. These metallized TMV-templated particles are predicted to play an important role in the future of nanodevice wiring.
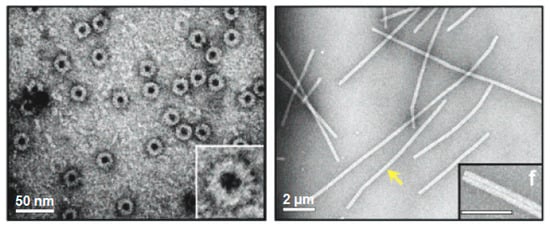
Another interesting application of TMV has been in the creation of light-harvesting systems through self-assembly. Recombinant coat proteins were produced by attaching fluorescent chromophores to mutated cysteine residues. Under appropriate buffer conditions, self-assembly of the modified capsids took place forming disc and rod-shaped arrays of regularly spaced chromophores (
Figure 3). Due to the stability of the coat protein scaffold coupled with optimal separation between each chromophore, this system offers efficient energy transfer with minimal energy loss by quenching. Analysis through fluorescence spectroscopy revealed that energy transfer was 90% efficient and occurs from multiple donor chromophores to a single receptor over a wide range of wavelengths
[69]. A similar study used recombinant TMV coat protein to selectively incorporate either Zn-coordinated or free porphyrin derivatives within the capsid. These systems also demonstrated efficient light-harvesting and energy transfer capabilities
[70]. It is hypothesized that these artificial light harvesting systems can be used for the construction of photovoltaic and photocatalytic devices.
Figure 3. Viral protein-based nanodisks and nanotubes. TEM images of chromophore containing nanodisks (
left) and nanotubes (
right) produced from a modified tobacco mosaic virus (TMV) coat protein
[69]. The scale bars represent 50 nm (
left) and 200 nm (
right). The yellow arrow is pointing to a single 900-nm-long TMV PNT containing over 6300 chromophore molecules. (Reprinted with permission from Miller et al.
J. Am. Chem. Soc.
129, 3104-3019 (2007)
[69]).
3.2. Cowpea Mosaic Virus (CPMV)
The cowpea mosaic virus (CPMV) is approximately 30 nm in diameter with a capsid composed of 60 copies of both large (L, 41 kDa) and small (S, 24 kDa) proteins
[71]. This icosahedral virus has coat proteins with exposed N- and C-termini allowing for peptides to be added onto the surface through genetic engineering. For example, virus-templated silica nanoparticles were produced through attachment of a short peptide on the surface exposed βB-βC loop of the S protein
[72]. This site has been most frequently used for the insertion of foreign peptides between Ala22 and Pro23
[73]. CPMV has also been widely used in the field of nanomedicine through a variety of in vivo studies. For example, it was discovered that wild-type CPMV labelled with various fluorescent dyes are taken up by vascular endothelial cells allowing for intravital visualization of vasculature and blood flow in living mice and chick embryos
[74]. Moreover, the intravital imaging of tumors continues to be challenging due to the low availability of specific and sensitive agents showing in vivo compatibility. Brunel and colleagues
[75] used CPMV as a biosensor for the detection of tumor cells expressing vascular endothelial growth factor receptor-1 (VEGFR-1), which is expressed in a variety of cancer cells including breast cancers, gastric cancers, and schwannomas. Therefore, a VEGFR-1 specific F56f peptide and a fluorophore were chemically ligated to surface exposed lysines on CPMV. This multivalent CPMV nanoparticle was used to successfully recognize VEGFR-1-expressing tumor xenografts in mice
[75]. In addition, use of the CPMV virus as a vaccine has been explored by the insertion of epitopes at the same surface exposed βB-βC loop of the small protein capsid mentioned earlier. One group found that insertion of a peptide derived from the VP2 coat protein of canine parvovirus (CPV) into the small CPMV capsid was able to confer protection in dogs vaccinated with the recombinant plant virus. It was found that all immunized dogs successfully produced increased amounts of antibodies specific to VP2 recognition
[76].
3.3. M13 Bacteriophage
The M13 bacteriophage is perhaps the most widely studied virus in terms of bionanotechnology and nanomedicine. The virion is approximately 6.5 nm in diameter and 950 nm in length enclosing a circular single-stranded DNA genome. The helical capsid is composed of approximately 2700 copies of the major pVIII coat protein and is capped with 5 copies each of the pIII, pVI, pVII, and pIX minor coat proteins
[77]. The process of phage display, which utilizes the ease of genetic manipulation to modify the surface proteins the M13 phage
[78], has enabled this simple phage to be used for multiple purposes including peptide mapping
[79], antigen presentation
[80][81], as well as a therapeutic carrier and bioconjugation scaffold
[82].
Recently, the major capsid protein of the M13 virus has been genetically engineered to display substrate binding peptides on the outer surface to selectively bind various conducting molecules
[83]. For example, recombinant pIII and pVIII coat proteins were used to select for peptide motifs that facilitated the formation of gold nanowires. Through an affinity selection/ biopanning process, a strong gold binding motif on pVIII containing four serine residues was identified
[77], a motif shown to have a high affinity for gold lattices
[84]. A streptavidin-binding 12-mer peptide was also inserted into the pIII coat protein for localization at one end of the helical capsid. Incubation with pre-synthesized 5-nm gold nanoparticles produced an ordered arrangement of the particles along the virion surface. The resulting Au-plated nanowires reached dimensions of 10 nm in diameter and approximately 1 µm in length
[77]. Similarly, Nam and colleagues developed negative electrodes for use in lithium ion batteries using highly ordered M13-templated gold-cobalt oxide nanowires
[85]. To do this, the group engineered a modified pVIII coat protein containing four consecutive N-terminal glutamate residues to bind cobalt oxide (Co
3O
4) along with an additional gold-binding peptide motif. This hybrid clone expressing both Au- and Co
3O
4-specific peptides produced a nanowire consisting of a small amount of Au nanoparticles combined with Co
3O
4. The hybrid nanowire was observed to improve initial and reversible storage capacity by approximately 30% compared to pure Co
3O
4 nanowires when tested at the same current
[85]. In a later study
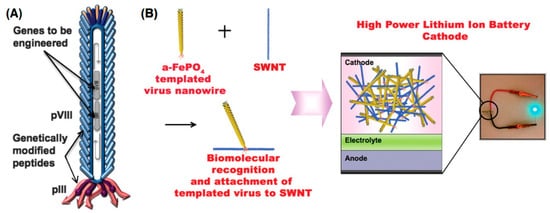
[86], the pVIII protein was bound to FePO
4 while the pIII protein was modified with a peptide sequence facilitating the interaction with single-walled carbon nanotubes (SWCNTs). This brought together the benefits of biologically ordered nanowires with the robustness of carbon nanotubes to produce high-power lithium-ion battery-like cathodes (
Figure 4)
[86].
Figure 4. Genetically engineered M13 bacteriophage used as a lithium-ion battery cathode. (
A) The gene VIII protein (pVIII), a major capsid protein of the virus, is modified to serve as a template for amorphous anhydrous iron phosphate (a-FePO
4) growth. The gene III protein (pIII) is also engineered to have a binding affinity for single-walled nanotubes (SWNTs). (
B) The fabrication of genetically engineered high-power lithium-ion battery cathodes and a photograph of the battery used to power a green light-emitting diode (LED). (Reprinted with permission from Lee et al.
Science 324, 1051–1055 (2009)
[86]).
Similar to CPMV, the M13 bacteriophage has been explored for use in cancer cell imaging and targeted drug delivery. Chemical modification of reactive groups on the M13 bacteriophage allowed for the attachment of small fluorescent molecules along with folic acid along its surface. Folic acid binds to the folate receptor, which is overexpressed in several cancers, facilitating uptake by the cell through endocytosis. The study found that successful binding and uptake of the dually modified bacteriophage by human BK cancer cells, enabling a multi-modal imaging platform
[87].
In addition, the M13 bacteriophage has been shown to penetrate the central nervous system (CNS), which has made it the focus of studies looking to deliver protein antibodies across the blood–brain barrier. The first example utilizing the M13 phage as a vehicle for transporting surface-displayed antibodies to the CNS was undertaken for the early detection of Alzheimer’s disease
[88]. In Alzheimer’s, characterized by the formation of β amyloid peptide (AβP) plaques, early detection is critical to obtain maximum benefits from available treatments. While there are many methods to detect amyloid plaques in post-mortem brain tissue, an effective in vivo imaging method remains elusive. A β-amyloid antibody fragment for specific detection of plaques in transgenic mice was used while for construction of a single-chain variable fragment (scFv), variable regions of the heavy and light genes of parental anti-AβP IgM 508 antibody were used
[73]. The resulting scFv-508F fragment was fused to the minor coat protein pIII and the recombinant phage successfully delivered phage-displayed anti-β-amyloid antibodies into the brains of mice via intranasal administration
[88]. Subsequent studies performed with radiolabeled antibodies containing an isotope suitable for in vivo diagnostic imaging (e.g.,
123I) suggests that this approach could allow for early detection of the disease
[89]. Similar research has looked at using antibody-displaying bacteriophage constructs for the treatment of drug addictions such as cocaine
[90]. Other protein-based approaches, such as the use of catalytic antibodies specific for the cleavage of cocaine, have not been successful in crossing the blood–brain barrier. Therefore, the pVIII coat protein containing a phage-displayed murine monoclonal antibody termed GNC 92H2 with high affinity and specificity for cocaine were assembled and administered to rats with no observed physical side effects. Enzyme-linked immunosorbent assay (ELISA) analysis of rat serum from vaccinated subjects showed no appreciable production of antibodies to the phage, demonstrating that an immune response was not occurring
[90]. These studies reveal that recombinant M13 bacteriophage offers a unique strategy to introduce therapeutic protein agents directly to the CNS.