All the natural CRSIPR/Cas networks are composed of many
Cas genes, which are encoded by homologous palindromic repeated units, RNA-mediated endonucleases, and novel short RNAs, termed as “spacers” produced by the introduction of short mobile sequences called protospacers. The protospacers are derived from the unique spacers which move among the homologous palindromic sequences repeats when the cell is attacked by invaders. These spacers work as identifying units for the invaded cell and allow the CRISPR-Cas system to cut foreign DNA sequences. There are three steps to the CRISPR/Cas-mediated immune system, including adaptation, expression, and interference. The first step in this mechanism is adaptation, which is associated with the sequential layout of the CRISPR-array via a new spacer’s procurement. Precursor CRISPR-RNA (pre-crRNA) and
Cas genes are expressed in the expression stage. Mature cr-RNAs are produced from precursor CRISPR-RNA using RNase III and Cas proteins in the interference event-specific targeted portion memorized by the combinative properties of both Cas proteins and cr-RNA
[17]. The CRISPR motif, termed as protospacer adjacent motif (PAM), is connected with each protospacer and closely situated in the target portion of the sequence. The PAM was found to be a highly specific part of the foreign phage or virus genome but is not present on the CRISPR locus in bacterial genome
[12].
The PAM sequence consisting of conserved dinucleotides is required upstream of the binding sites of crRNA for Cas proteins to recognize the target sequence
[18]. The Cas proteins are unable to detect target DNA for effective cleavage during PAM site recognition. The PAM is also exceptionally crucial to distinguish between the bacteria’s own DNA and invader DNA sequences. Such features enable bacteria to defend their own DNA from nucleases
[19]. In several kinds of CRISPR networks, PAM sequences are essential for Cas proteins functions, such as PAM sequence 5′-NNNNGATT which is targeted by Cas proteins in
Neissseria meningiditis [20]. Similarly, Cas9 proteins target the PAM sequence 5′-NGGNG or 5′-NNAGAA in
S. thermophiles [13][21] and 5′-NGG in
S. pyogenes [18].
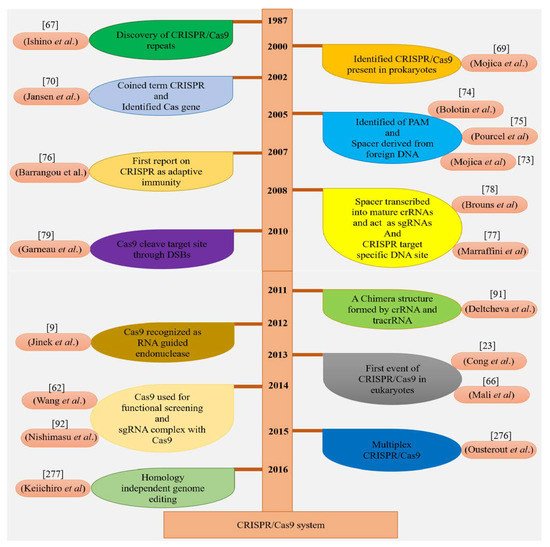
Two independent groups of scientists discovered the CRISPR/Cas9 machinery with three major kinds (type I–II–III) and two classes in host cells
[22]. In 2015, Markarova and coworkers executed the comparative genomic analysis of existing data and found two further reputed types and five subtypes
[23]. During the defense mechanism of CRISPR/Cas9 against invader DNA, these two classes behave differently, such as class 1 which consists of subtypes (I, III, IV) and uses many Cas proteins, while only a large Cas protein is used by the class 2 system, which has the subtypes (II, V)
[24]. In the adaptation phase of the CRISPR/Cas mechanism, spacers are added by Cas1 and Cas2 proteins and pre-crRNA develop involving Cas5 or Cas6 in the type I system. The Cas6 protein is also used in the type III system for a similar process but stimulation of 3′ end is accomplished by an uncertain element. For crRNA maturation, trans-activating crRNA (tracrRNA) and RNase are utilized in the type II mechanism
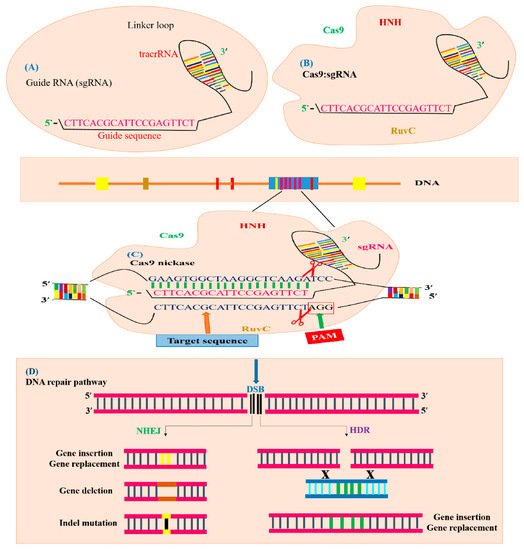
[25], as shown in (
Figure 2A). Currently, the immune system of
S. pyogenes operates as a type II system, which is a well-established GE technique known as CRISPR. This CRISPR/Cas9 system is modified by two major units: a non-coding chimeric RNA and Cas9 endonucleases for double-stranded (dsDNA) breaks in DNA (
Figure 2B)
[18]. The Cas9 protein is directed by the guide RNA (gRNA) and Cas9 proteins recognize targeted DNA in the presence of the “seed” sequence, which is produced by spacers derived from crRNA and the
S. pyogenses Cas9 (SpCas9) 5′ NGG ′3 sequence lying closely to the target region
[18]. The crRNA and tracrRNA are complementary to each other and it directs pre-crRNA to mature crRNA by means of RNase III. After the maturation of crRNA, it guides Cas9 proteins to break specific DNA sequences
[26]. In 2014, Nishimasu et al. (2014) demonstrated that the SpCas9 and gRNA DNA endonuclease has unique lobes, such as an assembly composed of a target detection lobe which is attached to the heteroduplex of sgRNA: a DNA molecule and a nuclease lobe which nicked the target DNA sequence
[27], as illustrated in (
Figure 2C).
Figure 2. Illustration of CRISPR/Cas9-mediated GE. The CRISPR/Cas9 system is composed of sgRNA and Cas9. (A) sgRNA with a guide sequence (colored pink) is developed by the combination of protospacer with crRNA and tracrRNA. (B) Cas9 machinery combines with sgRNA to form a complex to trigger CRISPR/Cas9 editing. The Cas9 nuclease consists of two parts, depending on its function and structure. The recognition site identifies the target DNA and interacts with sgRNA. The nuclease site contains two domains RuvC-like and HNH which cleave the target DNA site non-complementary by the RuvC domain and complementary by the HNH domain to the gRNA. (C) The Cas9 nuclease detects the genomic target site (indicated with blue color) having a 20 bp target sequence that is homologous to seed or guide sequence (indicated with pink color), which is crucial for Cas9 activity and specificity. The specific PAM sequence (indicated with red color) is detected by Cas9: sgRNA complex and DSBs created by the Cas9 endonuclease three base pairs upstream of the PAM sequence. (D) Targeted mutagenesis of a desired gene is achieved by filling the DSB (indicated with black color) by means of the HDR or NHEJ mechanism. The NHEJ repair mechanism generally produces insertion (indicated with yellow color), deletion (indicated with brown color) or indels (indicated with black line) at the break point, generating targeted mutants. The HDR repair mechanism uses a template DNA sequence for homologous recombination to produce gene replacement or gene insertion (indicated with green color).