Antimicrobial resistance (AMR) is a significant threat to global health. Antimicrobial peptides (AMPs) have shown potential as alternative diagnostic and therapeutic agents in biomedical applications. Their clinical applications are limited to topical application due to their systemic toxicity, susceptibility to protease degradation, short half-life, and rapid renal clearance. To circumvent these challenges and improve AMP’s efficacy, different approaches such as peptide chemical modifications and the development of AMP delivery systems have been employed. Nanomaterials have been shown to improve the activity of antimicrobial drugs by providing support and synergistic effect against pathogenic microbes.
1. Introduction
Antibiotics have been used for decades to cure infectious diseases and enabled most of modern medicine. Without antibiotics, even routine medical procedures can lead to life-threatening infections. The first widely used antibiotic, penicillin, was discovered in 1928. In 1945, Alexander Fleming warned that bacterial resistance had the potential to ruin the miracle of antibiotics
[1]. Shortly thereafter, beta-lactam antibiotics were discovered and proved to be effective against penicillin-resistant microbes
[2][3]. This was followed by the methicillin-resistant
Staphylococcus aureus (MRSA)
[4]; since then, resistance has been reported against almost all known antibiotics to date
[5]. In the high-priority list of antibiotic-resistant strains that require urgent attention are
Enterococcus faecium,
Staphylococcus aureus (
S. aureus),
Klebsiella pneumoniae (K. pneumoniae),
Acinetobacter baumannii,
Pseudomonas aeruginosa (
P. aeruginosa), and
Enterobacter species (ESKAPE). The ESKAPE are among the 12 microorganisms that are listed as critical to medium priority pathogens by the World Health Organization (WHO). These bacteria place a significant burden on the healthcare systems and global economic costs
[2][3]. Efforts to impede the spread of these pathogens have been hindered by their ability to resist antibacterial drugs.
As it stands, the misuse and overuse of these drugs are the major contributing factors toward antibiotic resistance, which often reduces the efficacy of newly discovered antibiotics and their derivatives
[6]. Antibiotic-resistant infections account for about 700,000 mortalities per year, which is estimated to increase to over 10 million deaths by 2050, making AMR a global health crisis
[7]. AMR occurs naturally, but the process has been accelerated by the misuse of antibiotics in both humans and animals. As reported by WHO, increased hospitalization, higher medical costs, and elevated mortality have been associated with AMR. AMR threatens the successful treatment of infections caused by drug-resistant microbes
[8]. Infectious diseases such as pneumonia, tuberculosis, gonorrhea, malaria, HIV/AIDS, and salmonellosis are becoming increasingly difficult to manage as known potent antimicrobial agents are becoming less effective. In this context, the loss of antimicrobial potency due to resistance in addition to the lack of new and alternative antimicrobial agents underscores the requirement for novel therapeutic agents.
AMPs of natural and artificial origin have gained attention in recent years due to their biological activities as potential alternatives to conventional antimicrobial agents that can combat pathogenic and drug-resistant microorganisms
[9]. These biomolecules serve as a natural first-line of defense system against invading pathogens, working either individually or synergistically with the innate immune system to inhibit the growth of pathogenic microorganisms. AMPs are divided into four categories based on their structural conformation and characteristics. This class of peptides (AMPs) are stable over a wide pH range, can work in synergy with immune cells, and possess effective antimicrobial activities against all kinds of pathogens including viruses. The discovery of defensins among other identified AMPs have been a major breakthrough as alternative molecules for use against antibiotic-resistance and in the development of novel antimicrobial agents
[10].
Over 14 manually curated AMP databases have been developed to provide information and enable researchers to synthesize AMPs with better therapeutic index. Examples of these databases include the Database of Antimicrobial Activity and Structure of Peptides (DBAASP v3.0)
[11], The Antimicrobial Peptide Database (APD3)
[12], dbAMP
[13], Yet Another Database of Antimicrobial Peptides (YADAMP), etc. To give insight into the growing number of available AMPs, the APD3 for example is composed of 3257 AMPs from six kingdoms (bacteria, archaea, protists, fungi, plants, and animals) with a broad spectrum of antimicrobial activities
[14].
AMPs have been used in the treatment of microbial infections emerging from bacteria, fungi, and viruses
[15]. The two most studied AMPs of human origin are the cathelicidin LL-37 and defensins. The antiviral effect of defensins has been demonstrated against human viral infections
[16][17][18]. The recombinant human β-defensins (mouse β-defensin 3) showed antiviral activity against influenza A virus in both in vitro and in vivo studies
[17]. Several studies have also demonstrated the immunomodulatory
[19][20], antimicrobial
[21], and wound-healing activities of the human LL-37
[22]. Of note, LL-37 interacts with keratinocytes through the P2X7–SFK–Akt–CREB/ATF1 signaling
[23][24] and COX-2
[25] pathways as well as with fibroblasts through the P2X7R pathway
[26] and the protein kinase/ERK pathway, supporting its healing effect in polymicrobial-infected wounds
[27][28].
2. Antimicrobial Agents and their Activity
The correlation between microorganisms and infectious diseases is well established. Therefore, molecules that can kill, inhibit, or slow down the growth of these pathogens are vital for treatment of microbial infections. After the discovery of
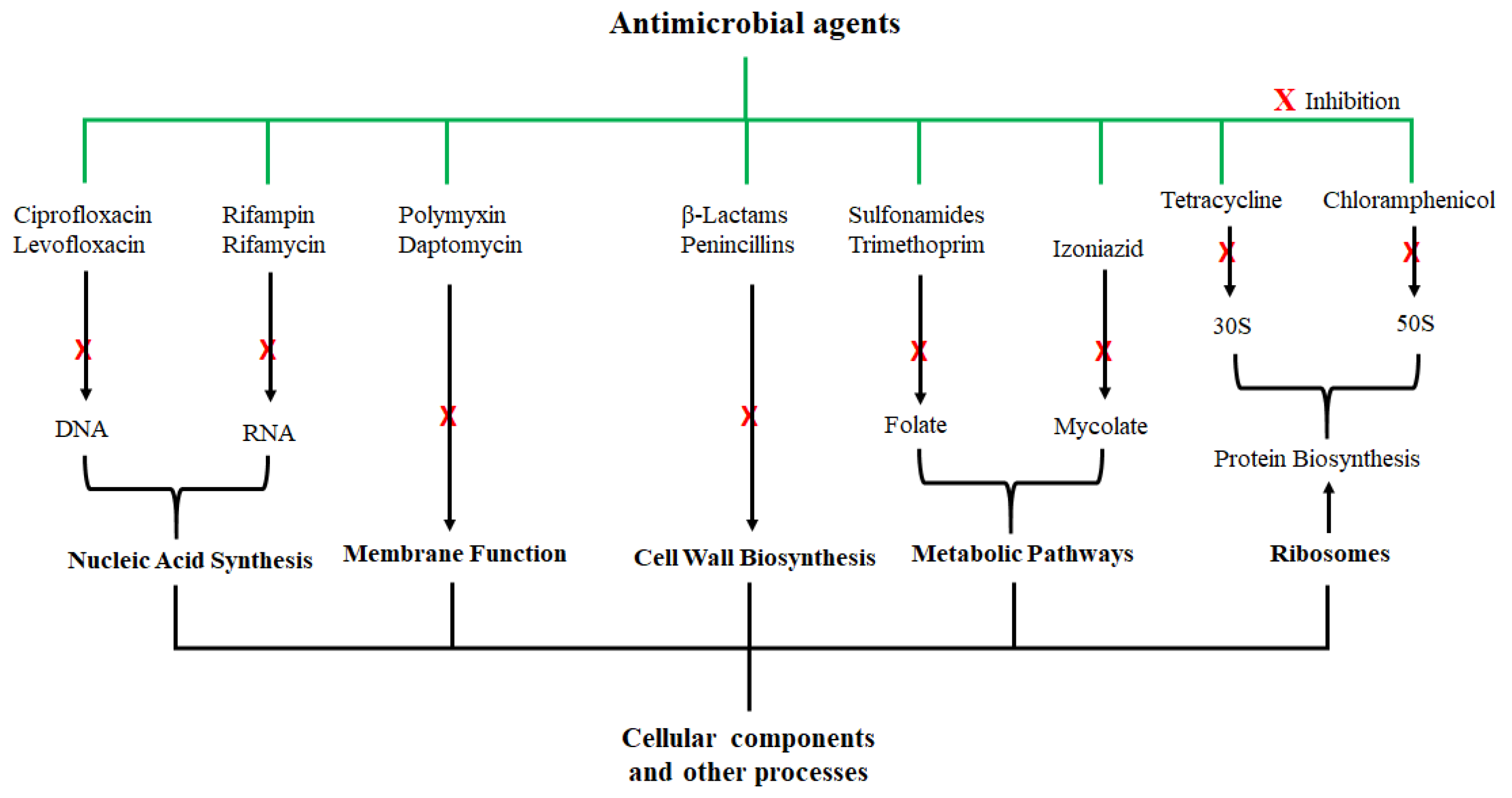
Penicillium notatum by Sir Alexander Fleming in 1928, several other antimicrobial agents were identified, and their mechanisms of antimicrobial activity have also been thoroughly investigated. Some of the antimicrobial agents and their modes of action are shown in
Figure 1; they exert their antimicrobial actions by interfering with various cellular and metabolic processes of the microorganisms. Actions of antibacterial agents can either be bacteriostatic if the antimicrobial activity involves growth inhibition or bactericidal if the activity involves membrane disruption leading to death of the bacteria
[29]. Sulfonamide and spectinomycin inhibit folate and protein synthesis, respectively; and are classified as bacteriostatic agents. Antibacterial agents such as vancomycin and penicillin are bactericidal agents due to their killing effect on bacteria. Other classifications of antibacterial agents are based on the mechanism of inhibition, origin, composition, and spectrum activity
[30]. While these agents were initially considered highly potent against certain microorganisms, the development of microbial resistance has been reported for nearly all these antibiotics. To make matters worse, the rate of discovering new antimicrobial agents has also been declining.
Figure 1. Examples of antimicrobial agents and their modes of action. These compounds are classified according to their cellular or molecular targets.
The misuse and overuse of antibiotics is the main cause of AMR, and have led to the increasing resistance of human and animal pathogens to these antimicrobial agents. The mode of microbial resistance toward the antimicrobial agents include (a) alteration or modification of the drug, the drug target, and drug binding, (b) inactivation of drugs, (c) blockage or decrease in drug uptake or penetration, (d) increase in drug efflux, and (e) the degradation of the drug. In 2015, 8.7% and 24.3% AMR were reported for MRSA and
Streptococcus pneumoniae, respectively
[31], and the mechanism of bacterial resistance was through drug inactivation, increased efflux, and ribosomal protection
[32].
3. Overview and Properties of AMPs
AMPs, also known as host defense peptides, are short amino acid sequences or biomolecules, ranging from 12 to 100 amino acids in length. AMPs possess antimicrobial activities against various microorganisms and are crucial to both the innate and the acquired immune systems as a defense mechanism
[33]. Some of these AMPs have also been shown to have antimicrobial activities against multidrug-resistant (MDR) bacterial strains. They are cationic and amphiphilic in nature
[34], and these characteristics play a major role in the mechanism by which they intercalate with the phospholipid bilayer of the microbial cell membrane, leading to membrane depolarization and cell permeabilization
[35]. Consequently, this will cause the release of biologically important cellular contents and subsequently result in microbial death
[36][37].
AMPs are classified based on their sequence composition, structure, and origin; typical AMP structural conformations are highlighted in
Figure 2 and
Table 1. The structural and physicochemical properties of AMPs, which include charge, hydrophobicity, and amphipathicity, dictate their specificity against the target microorganisms
[38].
Figure 2. Structural conformations of AMPs. (
A) α-helical, (
B) β-sheet, (
C) αβ-peptides, and (
D). Non-αβ peptides or extended structure. Images were deciphered by Schrodinger software v2020-3 after structural retrieval from the Protein Data Bank (PDB) at
https://www.rcsb.org/ (accessed on 20 May 2021) using the PDB IDs: 2K6O, 1ZMM, 1FD3, and 1G89 for A to D, respectively. AMPs are colored by properties.
Table 1. Classifications of some AMPs with specific examples.
|
Groups
|
Characteristics
|
Examples
|
Mode of Action
|
Refs
|
|
α-helical peptides
|
Amidated C-terminus,
N-terminal signal peptides
|
FALL-39
Magainins Cecropins
|
Pore formation
|
[39]
|
|
[40][41]
|
|
[42]
|
|
β-sheet
|
cationic with disulfide bridges
|
β-defensins
|
Membrane disruption
|
[43][44]
|
|
plectasin
|
[45]
|
|
protegrins
|
[46]
|
|
Extended AMPs or Non-αβ peptides
|
Contains proline, arginine, tryptophan, glycine or histidine rich amino acids
|
Indolicidin
|
Membrane disruption
Disruption of intracellular function
|
[47]
|
|
Bactenecins
|
[48]
|
|
Histatins
|
[49]
|
|
Loop peptides
|
|
Dodecapeptides
Tachyplesins
Protigrin-1
Bactenecin-1
Ranalexin
Brevinin 1E
Lactoferricin
|
Disruption of bacterial membrane
|
[50]
[51][52]
[53]
|
The antimicrobial activity of these peptides appears to be more rapid when compared to conventional antimicrobial drugs
[54][55], which together with new drugs are faced with the challenge of AMR. In light of this, there is a likelihood of AMPs succeeding where the conventional antimicrobial agents have failed and can possibly overcome microbial resistance due to their unique target interactions. AMPs execute their antimicrobial activities through attacks on the microbial membrane and as such would most likely require restructuring in the microbial membrane to bring about resistance to AMPs
[56]. The mechanism of AMP internalization is largely dependent on the molecular properties and membrane composition. Binding and interaction occurs through the electrostatic force of attraction between the negatively charged bacterial membrane and the positively charged amino acids of the AMPs, which is followed by hydrophobic interactions between the amphipathic AMP domains and the phospholipids in the microbial membrane
[54]. However, challenges such as proteolytic degradation, tissue toxicity, low stability, and difficulties associated with up-scaling must be met for AMPs to be considered as potential alternative to conventional antimicrobial drugs
[57]. Modified AMPs in combination with drug delivery systems can lead to the development of novel antimicrobial agents with improved therapeutic efficacy against MDR microbes
[58][59].
4. NPs with Antimicrobial Activity and Their Mode of Action
The search for novel antimicrobial agents has increased geometrically due to an increased incidence of microbial infections and AMR
[60]. Research advancement has also led to an increased use of NPs for biomedical applications due to their broad spectrum of activities against microorganisms. In vitro studies have shown the bactericidal activity of various nanomaterials against both Gram-positive and Gram-negative bacteria
[61][62][63], and similar findings were made using in vivo studies in mouse models that were infected with bacteria
[64].
The antimicrobial activity of NPs has been reported for various MNPs such as zinc oxide NPs, AuNPs, and AgNPs against a number of Gram-positive and Gram-negative bacteria
[65], fungi
[66][67][68][69], and viruses
[70][71][72]. Although most of the known polymeric NPs are commonly used as drug carriers, NPs fabricated using these polymers have been shown to possess endogenous activities. This applies to both passive (PLGA) as well as those with known antimicrobial activity (chitosan). Drugs loaded on these systems had improved biocompatibility and bio-activity. Drug-loaded PLGA-NPs exhibited higher antibacterial activity compared to the drugs alone or the unloaded NPs
[73][74]. Chitosan NPs enhanced the delivery and efficacy of HIV-1 P24 protein-derived peptides
[75]. Triclabendazole, which is used for the treatment of fascioliasis, is poorly soluble in water. The incorporation of this drug in chitosan NPs increased its bioavailability and stability at both low (pH 1.2) and high (pH 7.4) pH. The cytotoxic effects of the drug were significantly reduced in these nanoformulations. Thus, these nanosystems have the potential to lead to the development of orally ingested formulations for the treatment of fascioliasis
[76]. This section will emphasize on MNPs with antimicrobial activity and their mechanism of inhibition.
MNPs, which include AgNPs, AuNPs, and copper oxide NPs (CuO-NPs), have all been investigated and confirmed to possess antimicrobial activity against bacteria, viruses, and fungi
[77][78][79]. Although the exact mode of action of these NPs remains unclear, mechanisms such as reactive oxygen species (ROS) generation, metal ion release, and electrostatic interaction between the MNPs and the bacterial cell membrane have all been postulated. When compared to their respective salts, MNPs possess significantly higher antimicrobial activities against MDR microbes
[80]. The toxicity or antimicrobial effects of AgNPs
[81] and CuO-NPs
[82] on microbes was size-dependent, suggesting its role in the mechanism of microbial killing. The CuO-NP size was directly proportional to their antimicrobial activity, and also small-sized monodispersed NPs showed a significant increase in antibacterial activity
[82].
Specifically, the antimicrobial effect of AgNPs has been extensively studied against several microorganisms, and these are arguably some of the most promising MNPs used for the treatment of bacterial infections
[83]. Characteristics such as shape, size distribution, stability, and charge make them one of the most widely explored MNPs in science, medicine, and physical science
[84][85].
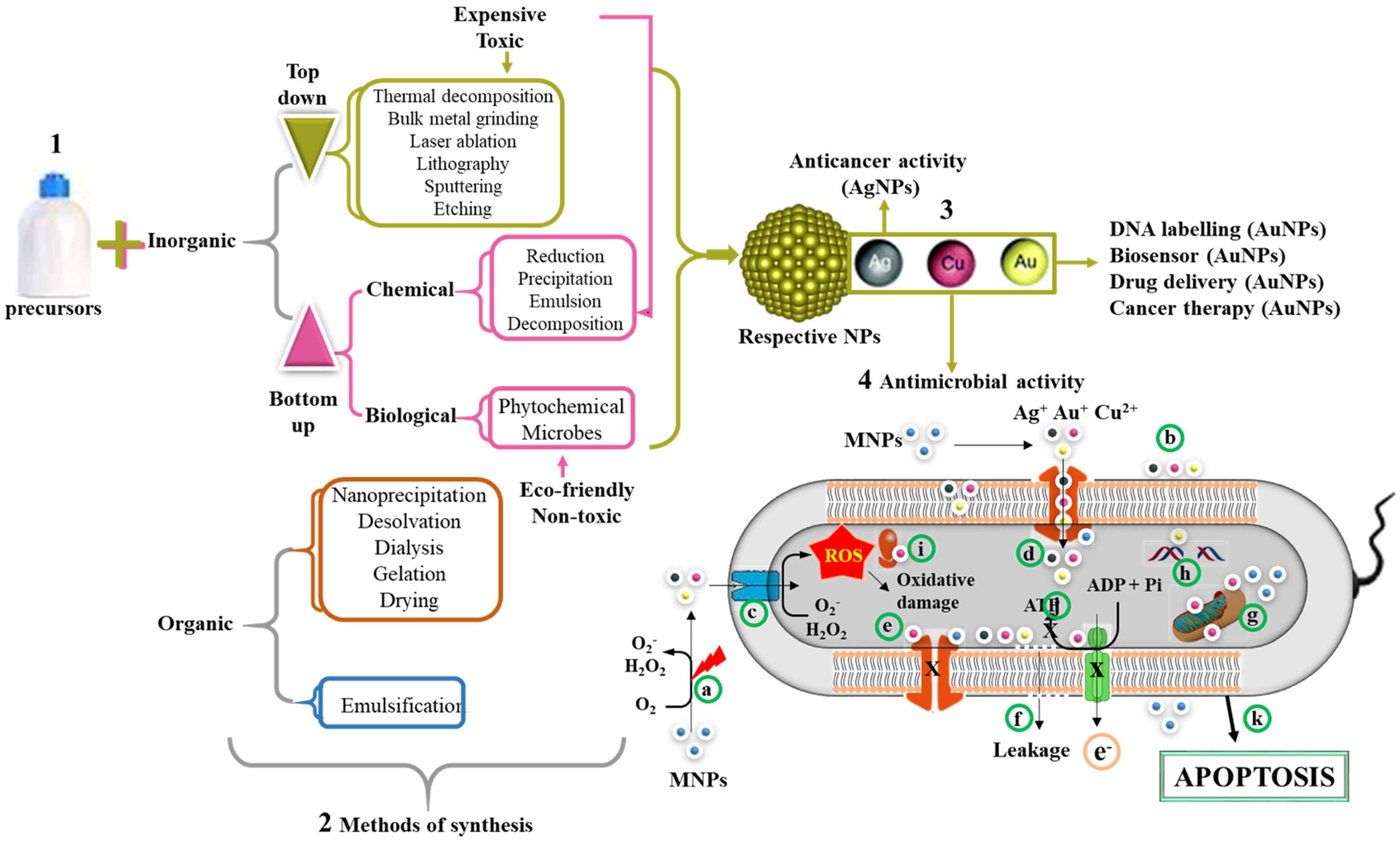
Figure 3 summarizes the mechanisms used in the synthesis of NPs, of which the chemical reduction method under the bottom–up approach is widely preferred over the top–down approaches. In recent years, green synthesis has been adopted to replace the toxic chemical reducing agents by benign natural resources such as microorganisms and plant extracts
[86]. This method is safer and eco-friendly when compared to physical and chemical methods of synthesis. The plant-mediated synthesis is economical and make use of readily available and renewable plant materials such leaves, stems, roots, etc. Moreover, synthesis occurs in just one step, since the phytochemicals are able to act as reducing, capping, and stabilizing agents
[86][87][88]. Previous studies have confirmed the safety of the biogenic method for MNP synthesis with effective antimicrobial activities against MDR bacteria
[88][89].
Figure 3. Synthesis of organic and inorganic NPs and their antimicrobial mechanism. MNPs can be synthesized using either a bottom–up or top–down approach (1). The methods of NP synthesis are broadly classified into physical, chemical, and biological methods (2). The reduction of metallic salts by these methods leads to the formation of MNPs. Examples of MNPs include AgNPs, AuNPs, and CuONPs (3). Various applications of MNPs include DNA labeling, biosensor, drug delivery, anticancer, and antimicrobial properties (4). The possible mechanism of their antimicrobial activity involves release of metallic ion when excited by laser (a) or in the presence of oxygen. The size of MNPs exhibited electronic effects and thus improved surface attraction (b). These metallic radicals in addition to their penetrative ability can easily pass through membrane channels (c). The MNPs triggers the generation of ROS inside the cells, which in turn leads to cellular damage. In addition, MNPs inhibit the channel transport of solutes/ions (e). The accumulation of these metallic ions leads to membrane depolarization and ultimately membrane leakage of cellular contents (f), mitochondrial dysfunction (g), DNA damage (h), ribosome disassembly (i), and inhibition of the electron transport chain and ATP synthesis (j). Collectively, all these processes can lead to cell death through apoptosis (k).
The antimicrobial property of
Acacia rigidula biosynthesized AgNPs was demonstrated against Gram-positive and Gram-negative MDR bacteria. The effect of the AgNPs was evaluated against
E. coli,
P. aeruginosa, and
Bacillus subtilis. Their safety profile was monitored in a murine skin infection model. The outcome of this study suggested that the AgNPs are compatible for use as a therapeutic agent against infectious diseases associated with drug resistant and drug susceptible bacterial strains
[74][90]. Due to their larger surface area, the surface of the MNPs can be modified to assign a specific activity by changing their surface composition. Lopez-Abarrategui et al. demonstrated that modification of citrate-coated MnFe
2O
4-NPs with antifungal peptide (Cm-p5) enhanced their antifungal activity in comparison to the free peptide and unmodified NPs. Cm-p5-MnFe
2O
4 completely inhibited
Candida albicans (
C. albicans) growth with a MIC of 100 µg/mL, compared to that of citrate-coated MnFe
2O
4-NPs at 250 µg/mL. The NPs showed no antibacterial activities against
S. aureus and
E. coli [91]. Therefore, MNPs could serve as excellent drug carriers due to their low cytotoxicity, ease of preparation, the ability to modify their surface, and good stability.
5. Nanocarriers of AMPs
NP–protein interaction possesses crucial application in biomedicine such as delivery systems and theranostic agents
[92]. However, the mechanism of recognition, specificity, and selectivity are poorly understood and remain a challenge. This section discusses the nanomaterials capable of transporting AMPs into the site of action in order to overcome their afore-mentioned limitations and exact the expected therapeutic effect.
Studies have reported specific microbial resistance and their mechanisms against AMPs
[93][94][95][96][97][98][99]. Pathogens can rapidly evolve and confer resistance to AMPs in vitro
[100]. Resistance evolution by Baydaa et al. was arguably the first study to explore the pharmacodynamic and bacterial AMP resistance. The study showed that AMP resistance in
S. aureus and some strains resulted not only in increased MICs but also an altered Hill coefficient (κ), resulting in steeper pharmacodynamic curves
[101]. Although AMR is also observed in AMPs, MDR against AMPs is not as prevalent when compared to antibiotics
[102]. Several approaches have been studied to improve the therapeutic use of AMPs. These include the combination of AMPs with traditional antibiotics since both have shown synergistic effect in the reduction of microbial resistance. Another method is the use of nanocarriers, which have been shown to reduce other side effects while exacting maximum suicidal activity against microbial populations
[103].
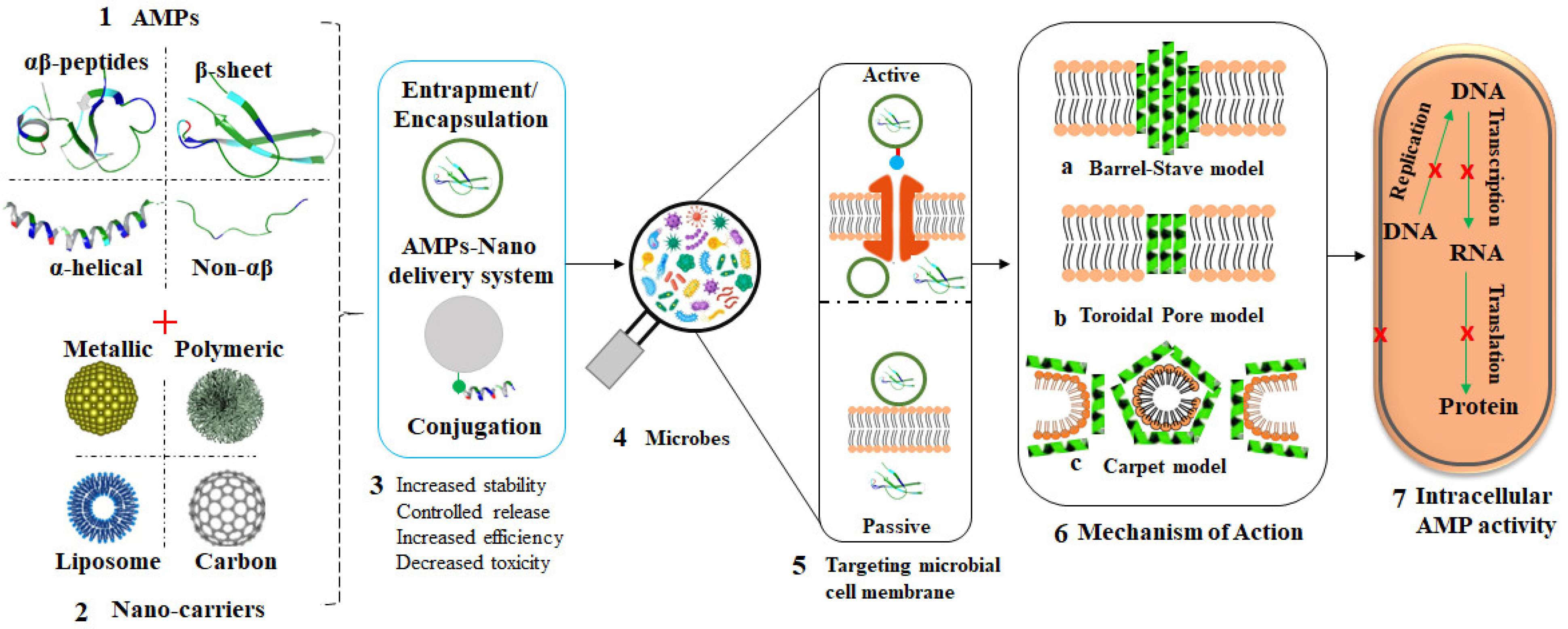
In a bid to prevent AMR or bypass drug resistance, AMPs are now being considered as potential alternative for antibiotics. Nanomaterials, especially polymeric and MNPs, provide one of the promising drug delivery systems, as highlighted in
Figure 4 [104][105][106]. Aside from the fact that some nanomaterials possess antimicrobial activities and can inhibit the growth of microbes through several mechanisms, they can also act as carriers for either antibiotics or AMPs to overcome the defense mechanisms of microbes and further enhance antimicrobial effects
[107]. These materials can be functionalized with antimicrobial agents to prevent and treat microbial infections as well as to improve the effectiveness of the conventional drugs
[108].
Figure 4. Overview of AMP-loaded nanocarriers and their mode of action. Structurally, AMPs are classified into four groups (1); different nanocarriers have been studied as effective carries of AMPs (2); different AMP nanoformulations can be obtained through various chemistries between peptides and nanocarriers (3); exposure of microbes to these AMP nanoformulations (4); via passive or active transportation (5); leading to bacterial membrane attack by the AMPs through various AMP-dependent mechanisms (6) and ultimately bacterial death (7).
Nanomaterials such as MNPs (AuNPs, AgNPs), polymeric NPs, dendrimers, liposomes, micelles, and carbon nanotubes have all been used as carriers or transporters of drugs
[109][110][111][112]. The function of these carriers is to decrease side effects, lower drug dosage, maintain constant drug levels in the blood, maximize therapeutic index, and reduce drug degradation and undesirable side effects
[113]. MNPs display striking different size and shape-dependent properties when compared to their bulk material. These properties include a wide surface plasmon resonance (SPR) band, which is directly correlated to particle size, a large surface to volume ratio, biocompatibility, and low toxicity
[114][115]. The MNPs’ parameters such as charge, size, and surface composition have all been reported to influence the activity of the NPs
[116][117][118]. MNPs have been explored and utilized in several biomedical applications such as therapy, drug and gene delivery, probes, sensors, diagnostics, and photocatalyst.
MNPs have been shown to improve the antimicrobial activity of drugs by providing support and synergistic effects against pathogenic microbes. Specifically, MNPs have incredible physicochemical properties and have shown novel bioactivities, which can be enhanced by attaching bioactive molecules
[119]. The functionalization of MNPs is achieved by conjugating different molecules through different mechanisms. MNPs employed in biological applications are usually modified with biocompatible polymers such as polyethylene glycol (PEG), proteins/peptides (e.g., bovine serum albumin), and oligonucleotides. The process of linking molecules to the surface of the MNPs can be achieved by physisorption or by taking advantage of the metal’s affinity for the sulfhydryl group of thiolated molecules. Electrostatic interactions and non-covalent conjugation can also be used to functionalize the MNPs with molecules that contain reactive groups such as hydroxyl, carboxylic group, and amine groups, which can be used to attach other biologically active molecules.
The biologically active moieties can include small drug molecules
[120] or proteins
[121], while targeted delivery/therapy can be achieved by conjugating target specific aptamers
[122], antigen/antibodies
[123], or peptides
[124]. Different conjugation chemistries have been studied for effective drug loading and delivery. Examples of these chemistries include the conjugation of RR-11a endopeptidase to liposomes using the amine/carboxyl chemistry
[125], the polyclonal Rabbit IgG conjugated to AuNPs using the amine/carboxylate chemistry
[126], S2P peptide (CRTLTVRKC) conjugated to chitosan NPs through the amine/carboxylate, thiol/maleimide chemistry
[127], liposome functionalized with adiponectin, globular domain by thiol/maleimide chemistry
[128], and the J18 RNA aptamer conjugated to AuNPs by the base-pairing hybridization
[129]. Immobilization of the biomolecules/AMPs on the NPs can occur in two ways; i.e., the AMPs are either temporarily (reversible) or permanently (irreversible) tethered on or within the NPs. The reversible chemistry ensures targeted delivery and release of the AMPs in its native form, and their functions are independent of the nanocarriers. The reversible nanosystems are usually responsive to biological stimuli, where a change in pH or presence of an analyte will trigger release of the attached AMPs. In the irreversible conjugation, the AMPs are permanently attached on the nanoparticle and act in synergy with the NPs
[130][131][132].
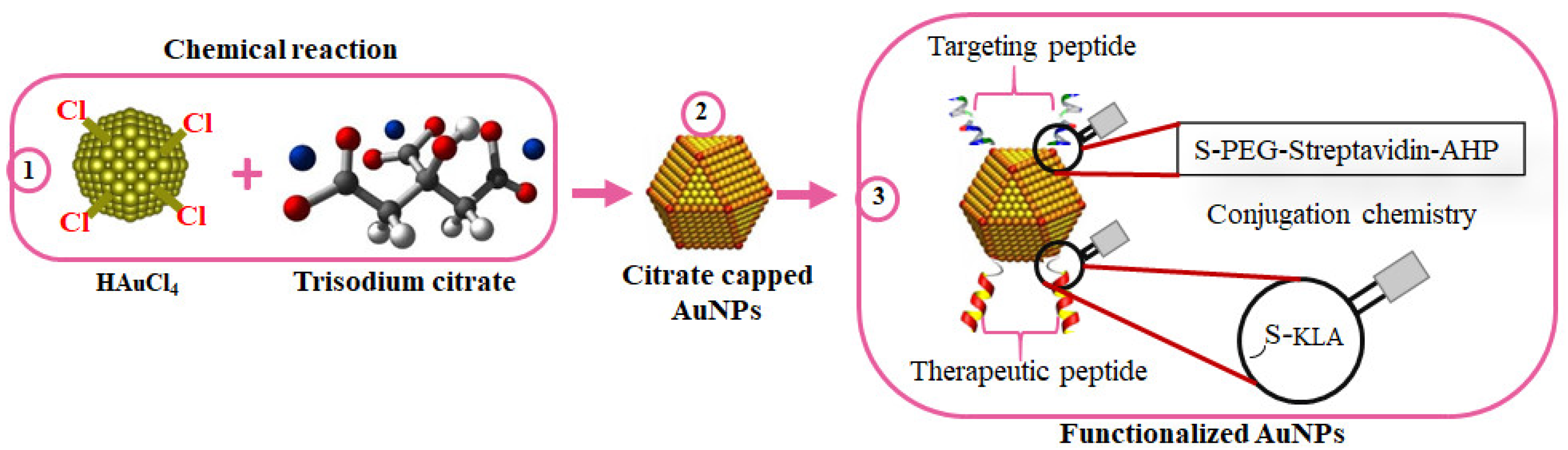
Several nanoconjugates with biologically active molecules have shown promising outcomes for the treatment of some diseases. In a comprehensive study by Sibuyi et al., AuNPs were used in the development of targeted nanotherapy for cancer. The AuNPs were bifunctionalized with adipose homing (targeting) moiety and an AMP (
DKLAKKLAK
2/KLA) with proapoptotic activity for the selective induction of apoptosis in target cells (
Figure 5). The homing peptide was conjugated to the AuNPs through an irreversible chemistry, while the KLA used a reversible chemistry where a caspase-3 cleavage site (DEVD) was used as a linker between the AuNPs and KLA. Upon internalization into the cells, the KLA is detached from the AuNPs by caspase-3 and triggers cell death through apoptosis
[133]. Similarly, KLA-loaded liposomes with the adipose homing peptide accumulated in the white adipose tissues (WATs), resulting in body weight loss in obese mice models, as shown by Hossen, et al.
[134]. The bifunctionalized NPs showed potential of repurposing AMPs for different applications by selectively targeted the cells that express the receptor for the targeting peptide, i.e., the colon cancer cells
[133] and the endothelial cells in the WATs of obese mice
[105].
Figure 5. Surface chemistry of bifunctionalized citrate-capped AuNPs. AuNPs were chemically synthesized using trisodium citrate as the reducing agent (1); to form citrate-capped AuNPs (2); while exploring the AuNP-thiol affinity for the attachment of the targeting and therapeutic peptides (3).