1000/1000
Hot
Most Recent

Extracellular vesicles (EVs) are mediators of communication by transferring messenger bioactive molecules including proteins, lipids, and miRNAs between cells and tissues. The specific functions of EVs principally depend on the internal cargo, which upon delivery to target cells trigger signal events that modulate cellular functions. The vesicular cargo is greatly influenced by genetic, pathological, and environmental factors.
Communication between cells is crucial to preserve body homeostasis and health. This is particularly relevant in the intestinal tract, where host cells are exposed to millions of bacteria and food antigens [1]. Besides cell-to-cell contacts and soluble released factors that can act locally or distantly on other cells types and tissues, extracellular vesicles (EVs) can mediate intercellular crosstalk. EVs are a heterogeneous population of lipid bilayer membrane-enclosed vesicles that transport and deliver proteins, lipids, and nucleic acids to recipient cells. There are multiple classes of EVs released from almost all cell types and they can also be found in various biological fluids. Initially, the release of EVs was considered a mechanism to discard unwanted materials from cells. However, intensive research has revealed that release of EVs has a pivotal role in cell-to-cell communication in human physiology and pathology.
The term EVs comprise a varied, heterogeneous group of particles released from cells that largely originate from endosomes and/or plasma membrane with complex cargoes [2]. The classification of EVs has been inconsistent and confusing. In fact, isolation methods for EV subtypes are still under development and consensus on best practices has not yet been reached. However, according to the International Society for Extracellular Vesicles (ISEV), minimal requirements must be met to assert the presence of EVs in sample isolates, and several experiments need to be conducted to characterize the existence of EVs [3]. Thereby, on the basis of current studies on the mechanism of formation, mode of release from cells, and size, we can describe three categories: (i) exosomes, (ii) microvesicles (MVs), and (iii) apoptotic bodies. Although vesicle size is the main factor used in EV categorization, some reports have demonstrated that EVs within one EV class may be heterogenic in terms of cargo and functionality.
Exosomes are small EVs defined as 30–200 nm lipid bilayer particles secreted by all cell types [4]. Recent studies have shown two novel subpopulations of exosomes, including large exosome vesicles (Exo-L) with 90–120 nm diameter, small exosomes vesicles (Exo-S) with 60–80 nm diameter, and an abundant population of non-membranous nanoparticles called exomeres with sizes around 35 nm [5]. Exosomes can be found in almost all living cells, including dendritic cells, lymphocytes, mast cells, intestinal epithelial cells, and endothelial cells. Moreover, exosomes can be detected and isolated from various body fluids such as urine, plasma, cerebrospinal fluid, human milk, and exudates [6]. Exosomes originate first as intraluminal vesicles (ILVs) within a multivesicular body (MVB), which are released into the extracellular matrix after MVB fusion with the plasma membrane. This process operates through the endosomal pathway [7]. Due to their origin, exosomes differ from other EVs that are released from the cell as the result of a direct budding process of the plasma membrane.
MVs, also called microparticles or ectosomes, are defined as 200–1000 nm vesicles released by vesiculation from eukaryotic cells [8]. They are formed by outward blebbing of the plasma membrane and subsequently released via actomyosin-driven fission of plasma membrane blebs. Initially, MVs were described as “platelet dust”, due to their identification as subcellular material originating from platelets in normal plasma and serum [9]. MVs derived from human cancer cells are called “oncosomes” (1–10 µm) and have a role in cell–cell communication. Their ability to participate in the horizontal transfer of signaling proteins and mediators contributes to invasive activity in cancer [10]. Although oncosomes are classified within MVs; this group is studied separately due to their larger size, characteristic biomolecules, and isolation methods.
Apoptotic bodies include much larger vesicles of 1–5 µm and they are formed during membrane blebbing and cellular disassembly from fragmentation when the cytoskeleton breaks at the beginning of apoptosis [11]. Their impact upon other cells is not well studied. Nonetheless, it has been suggested that apoptotic bodies participate in immune regulation, including processes such as autoimmunity, cancer, and infection [12]. This group will be excluded from this review.
It is known that EVs are formed by multiple mechanisms that are summarized in Figure 1. Exosomes are generated within the endosomal system and microvesicles are formed by outward budding vesicles at the plasma membrane. Although the generation of EVs (exosomes or microvesicles) occurs at distinct sites within the cell, common intracellular mechanisms are involved in the biogenesis of both entities.
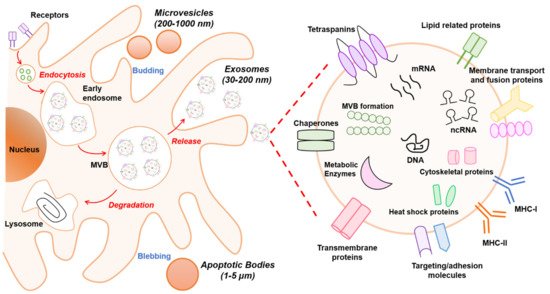
Figure 1. Schematic overview of extracellular vesicle (EV) biogenesis and cargo. At least three different subclasses of EVs are generated by eukaryotic cells: exosomes (30–200 nm), microvesicles (200–1000 nm), and apoptotic bodies (1–5 µm). The left panel schematically shows the biogenesis pathway for each EV type. Microvesicles and apoptotic bodies sprout directly from the plasma membrane, whereas exosomes are generated within multivesicular body (MVB) subpopulations that upon maturation fuse with the plasma membrane. The biogenesis pathway influences the cargo of EVs. In particular, the composition of exosomes is presented in the right panel. Exosomes are rich in the adhesion molecules tetraspanins (CD9, CD81, CD63), antigen-presenting molecules (MHCI/II), membrane transport proteins (annexins, flotillin), enzymes (elongation factors, metabolic enzymes), and other cytosolic proteins (ribosomal proteins). In addition, lipids (sphingomyelin and phosphatidylserine) and nucleic acids (DNA, RNA, non-coding RNAs (ncRNAs), and micro-RNAs (miRNAs)) also are bioactive components.
Exosomes are derived from endosomal compartments. Within the endosomal system, endosomes are divided into compartments including early endosomes, late endosomes, and recycling endosomes. To release exosomes, plasma membrane and cytosol-associated molecules (lipids, proteins, and nucleic acids) are endocytosed and transferred into early exosomes, which fuse with endocytic vesicles sorting their cargo for degradation, recycling, or secretion. The remaining early exosomes differentiate into late exosomes that give rise to ILVs formed by inward budding of the endosomal membrane. In turn, ILVs are enclosed within MVBs. These can either fuse with lysosomes if their content is destined for degradation, or merge with the cellular membrane, releasing the ILVs as exosomes into the extracellular space [13].
The exosome biogenesis pathway can be regulated by various mechanisms. One well-known mechanism involves the endosomal sorting complex required for transport (ESCRT) that is needed for MVB formation as it classifies ubiquitinated intracellular cargos that are destinated for lysosomal degradation into MVBs [14]. The ESCRT machinery consists of four multiprotein complexes (ESCRT 0, I, II, and III) and accessory proteins (TSG101, ALIX, Vps4, and VTA1). Although ESCRT units are released into the cytosol for recycling, some accessory proteins such as TSG101, HRS, and ALIX remain in exosomes as markers. Alternatively, recent evidence has shown that ESCRT-independent mechanisms exist for exosome formation and release that are based on neutral sphingomyelinase (nSMase)-dependent ceramide formation. This lipid could facilitate membrane invagination of ILVs through its cone-shaped structure [15][16]. Moreover, proteins from the tetraspanin family such as CD9, CD63, CD81, and CD82 are regulators of ESCRT-independent endosomal sorting. For instance, it has been reported that CD63 allows for sorting of a melanosomal protein in ILVs in human melanoma cells [17].
The mechanism involved in the secretion of EVs formed by direct budding from the plasma membrane, such as MVs, is less well characterized and has only started to emerge. Evidence has revealed that recruitment of the ESCRT-I subunits TSG101 and Vps4 to the plasma membrane through binding to a tetrapeptide protein within the Arrestin 1 domain-containing protein 1 (ARRDC1) results in the release of MVs containing TSG101, ARRDC1, and other cellular proteins [18]. Several factors such as redistribution of phospholipids including reposition of phosphatidylserine to the outer leaflet, contraction of actin/myosin machinery, and extracellular concentration of Ca+2 can impact MV formation and alter membrane fluidity and deformability [19]. In addition, enzymes that transfer lipids from one leaflet of the plasma membrane to the other such as flippases, floppases, and scramblases play an important role in MV development [20].
EVs are released to the extracellular space due to the presence of Rab proteins, which are essential regulators of intracellular vesicle transport between compartments. Rabs can be involved in vesicle budding, trafficking through interaction with the cytoskeleton, or docking to the membrane of an acceptor compartment [21]. Secreted EVs establish interactions with recipient cells and transmit the information. The interaction can be through ligand/receptor molecules on their respective surfaces mediated by classical adhesion molecules including integrin and intracellular adhesion molecules. Additionally, EVs can deliver their content by endocytic processes such as clathrin- and caveolin-mediated endocytosis, lipid raft endocytosis, phagocytosis, and micropinocytosis, probably guided by vesicle membrane composition and surface molecule profile [22].
The biochemical composition of EVs has been studied in several populations and likely depends on the mode of biogenesis, cell type, and physiological conditions. Likewise, the isolation methods that are used can influence the composition of EVs. Thus, characterization of all their components remains unclear. Recent technology advances and new methodologies such as proteomics, lipidomics, and transcriptomics have allowed the identification of different types of cargos in EVs (Figure 1). The derived information has been integrated in several databases such as Exocarta [23], Vesiclepedia [24], and EVpedia [25], which serve as repositories and tools that help to decipher vesicle loading and functions. Overall, all EVs are loaded with proteins, lipids, and nucleic acids, and their cargo can be specific to the vesicle and cell type. This section will describe the content of EVs in general terms.
Proteins: The protein cargo of EVs in various cell types has been extensively reviewed [26][27]. Diverse proteomics analyses of exosomes have identified proteins associated with the biogenesis mode, including proteins linked to the endosomal pathway [28]. In fact, components of ESCRT have been found in exosomes, including ALIX, TSG101, and HRS [29]. In addition, proteins involved in the trafficking and release of EVs such as RAB27A, RAB11B, and ARF6 are commonly identified [30]. As mentioned above, EVs are rich in tetraspanin proteins including CD63, CD81, and CD9, which are being used as exosome markers. These proteins can target specific cells, such as endothelial cells, to promote angiogenesis [31]. In addition, several metabolic enzymes such as ATPase, glyceraldehyde-3-phosphate dehydrogenase, enolase-1, and pyruvate kinase type M2 have been detected in EVs by proteomic analysis [32]. Likewise, heat shock proteins including HSP70 and HSP90 and major histocompatibility complex (MHC I and II) are found in EVs derived from most cell types and are involved in antigen binding and presentation [31].
Lipids: The content of lipids in EVs are not fully characterized, but in general it is known that lipid composition shares common features with the origin cells. Outstanding reviews about this topic are available in the literature [33][34]. It has been suggested that some lipids can be specifically associated with different types of EVs, depending on whether they originate from MVB or from plasma membrane [35]. In general, EVs are rich in phosphatidylcholine, phosphatidylserine, phosphatidylethanolamine, phosphatidylinositols, phosphatidic acid, cholesterol, ceramides, sphingomyelin, glycosphingolipids, and other less abundant lipids [36]. Exosomes are rich in phosphatidylserine facing the extracellular milieu, an attribute that likely facilitates their internalization by recipient cells [37]. A single universal marker for MVs is less well defined. However, ARF-6 and CD40 are frequently found in MVs derived from tumor cells [38]. Since EVs are membrane-limited vesicles, their lipid bilayer has a protective function that preserves internal cargo from proteolytic or RNAase degradation.
Nucleic acids: Recent research has demonstrated a varied composition of genetic material in EVs. Diverse studies have found genomic DNA, complementary DNA (cDNA), and transposable elements in extracellular vesicles [39]. Currently, research is focused on RNA content. Extensive scientific evidence shows that EVs carry functional RNAs that play a pivotal role in cell-to-cell communication and regulation of cellular processes. In this sense, EVs contain mRNAs, non-coding RNAs (ncRNAs) including micro-RNAs (miRNAs), small nuclear RNAs (snRNAs), and transfer RNAs (tRNAs) [40]. In particular, miRNAs exported through EVs have a relevant role since they can post-transcriptionally regulate gene expression in distant cells [41][42][43]. In this context, a database called EV miRNAs has been created with data from multiple studies analyzing miRNA expression profiles in various types of EVs from distinct cell types [44]. Since exosomal miRNAs play a pivotal role in several diseases, have great stability, and reach a high concentration in circulation, they have great potential as disease biomarkers for diagnosis or prognosis, and as therapeutic targets [8][45].
Intestinal homeostasis depends on complex, dynamic interactions between the microbiota, the epithelium, and the host immune system. Given the complexity of the intestinal ecosystem, regulatory mechanisms involving immune receptors, signaling pathways, regulatory proteins, and miRNAs are required to ensure symbiosis and avoid exacerbated inflammatory responses that could lead to pathological states. In this scenario, intercellular communication is crucial to orchestrate balanced responses to preserve intestinal homeostasis.
There is mounting evidence that intra- and interkingdom crosstalk is mediated by EVs released by gut microbiota or by host intestinal cells. This review is focused on host-derived EVs. It is well known that cargos in EVs confer specific properties and functions. For instance, the presence in EVs of metalloproteinases (MMPs), chemokines, cytokines, growth factors, adhesion molecules, and miRNAs plays a crucial role in processes involving the immune system. Differences in the amount of released EVs and their composition have been observed in health and disease states. High levels of EVs with different content were found in samples from patients with inflammatory bowel disease (IBD), which suggests their contribution to intestinal inflammatory processes [46].
The intestine is critical in controlling human health. Epithelial and immune cells of the intestinal mucosa are constantly exposed to millions of microbes that greatly influence the integrity of intestinal epithelial barrier and immune function. In this ecosystem, both inter-kingdom and intra-host–cell communications are key to control microbiota population and avoid exacerbated inflammatory responses. Host derived EVs have relevant role in mediating such interplay, controlling intestinal homeostasis at various levels, as summarized in Figure 2. Little is known about the function of host EVs in controlling gut microbiota, but recent reports point to faecal miRNAs secreted through EVs released by IECs to the gut lumen as modulators of this microbial community.
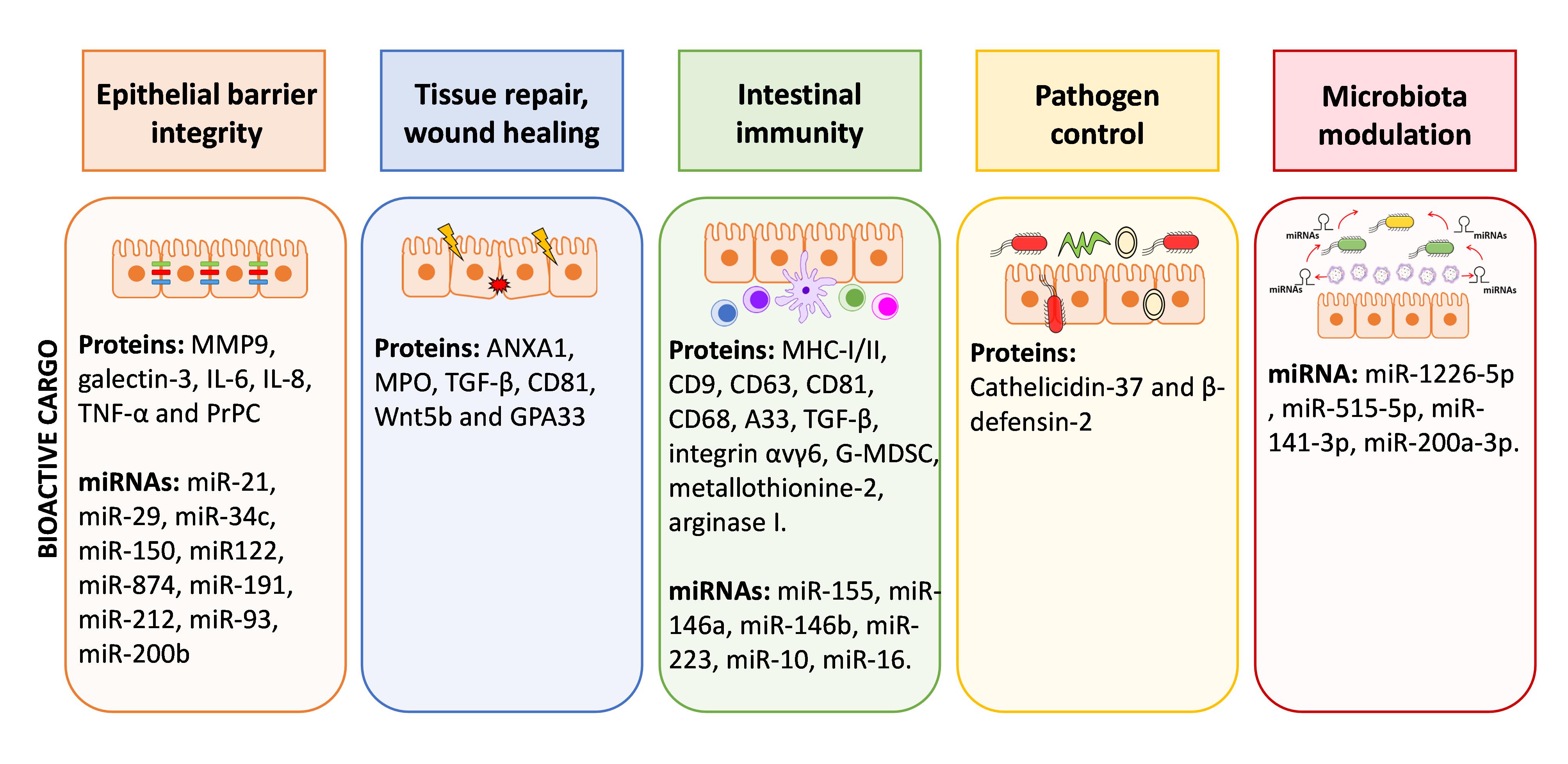
Figure 2. Graphical summary of functions of EVs within the gut environment. EVs derived from IECs and immune cells of the lamina propria contribute to cell-to-cell communication in the gut and have great impact on the homeostasis/inflammation balance. The scheme summarized the bioactive cargo in EVs that can influence epithelial barrier integrity, tissue repair, immune responses, control of pathogens and microbiota shaping.
The role of EVs as messengers in cell-to-cell communication inside the body is not limited to the gut environment. As mentioned above, it is now well accepted that circulating EVs can reflect diverse healthy and pathologic states of cells and tissues. Therefore, they are being explored as diagnostic biomarkers or therapy tools in several disorders.