1. Introduction
The Rho (Ras homologous) family of small GTPases is one of the members of the Ras superfamily. In humans, 20 Rho-family GTPase members have been identified and categorised into eight major subfamilies Rho-, Rac-, Cdc42-, RhoU/RhoV-(also known as Wrch/Wrch2), Rnd-, RhoD/RhoF-, RhoBTB- and RhoH subfamilies. The subfamilies are categorised based on the similarities in their primary amino acid sequence (), structural motifs and biological functions
[1]. Initially, Miro and RhoBTB3 were included as members of the Rho family small GTPases, but they have been excluded due to the lack of similarity between these proteins and other members of the Rho-family. For instance, Miro subfamily proteins lack the Rho insert region and are assumed to be non-catalytic as the sequence in the highly conserved G2–G5 loops responsible for nucleotide binding and hydrolysis is different compared with that in canonical GTPase domains
[2][3]. RhoBTB3 shares 25% sequence similarity with and contains the same BTB domains as RhoBTB1 and RhoBTB2. However, this protein is not considered as a Rho GTPase by some studies due to its lack of similarity with Rho and Ras
[3][4].
Table 1. The percentage sequence similarity shared between Rho-family GTPases.
| |
Rho
BTB1
|
Rho
BTB2
|
RhoH
|
Rnd1
|
Rnd2
|
Rnd3
|
RhoD
|
RhoF
|
RhoA
|
RhoC
|
RhoB
|
Wrch2
|
Wrch1
|
TC10
|
TCL
|
Cdc42
|
RhoG
|
Rac2
|
Rac1
|
Rac3
|
|
RhoBTB1
|
|
70
|
34
|
33
|
31
|
30
|
34
|
28
|
38
|
37
|
38
|
34
|
32
|
38
|
35
|
40
|
40
|
41
|
42
|
41
|
|
RhoBTB2
|
70
|
|
32
|
32
|
31
|
29
|
34
|
28
|
39
|
37
|
38
|
33
|
32
|
39
|
37
|
40
|
40
|
42
|
42
|
41
|
|
RhoH
|
34
|
32
|
|
29
|
32
|
36
|
38
|
33
|
40
|
40
|
41
|
41
|
38
|
40
|
39
|
42
|
40
|
40
|
41
|
40
|
|
Rnd1
|
32
|
32
|
29
|
|
53
|
61
|
37
|
39
|
41
|
42
|
41
|
31
|
32
|
36
|
34
|
37
|
37
|
39
|
39
|
38
|
|
Rnd2
|
31
|
31
|
32
|
53
|
|
63
|
39
|
41
|
46
|
47
|
43
|
28
|
31
|
36
|
35
|
37
|
41
|
40
|
41
|
39
|
|
Rnd3
|
29
|
29
|
36
|
61
|
63
|
|
37
|
40
|
48
|
48
|
47
|
31
|
32
|
39
|
35
|
38
|
41
|
39
|
42
|
40
|
|
RhoD
|
34
|
35
|
38
|
37
|
39
|
37
|
|
49
|
49
|
49
|
49
|
39
|
36
|
42
|
38
|
43
|
44
|
46
|
49
|
49
|
|
RhoF
|
28
|
28
|
33
|
39
|
41
|
40
|
49
|
|
47
|
48
|
47
|
36
|
37
|
46
|
43
|
43
|
46
|
50
|
59
|
47
|
|
RhoA
|
38
|
39
|
40
|
41
|
46
|
48
|
49
|
47
|
|
92
|
85
|
40
|
44
|
51
|
48
|
53
|
55
|
53
|
57
|
55
|
|
RhoC
|
37
|
37
|
40
|
42
|
47
|
48
|
49
|
48
|
92
|
|
85
|
40
|
44
|
50
|
49
|
51
|
55
|
53
|
57
|
54
|
|
RhoB
|
38
|
38
|
41
|
41
|
43
|
47
|
49
|
47
|
85
|
85
|
|
42
|
45
|
51
|
48
|
50
|
53
|
54
|
55
|
54
|
|
Wrch2
|
34
|
33
|
41
|
31
|
28
|
31
|
39
|
36
|
40
|
40
|
42
|
|
59
|
51
|
48
|
53
|
46
|
51
|
52
|
53
|
|
Wrch1
|
32
|
32
|
37
|
32
|
31
|
32
|
36
|
37
|
44
|
44
|
45
|
59
|
|
50
|
46
|
56
|
48
|
54
|
54
|
54
|
|
TC10
|
38
|
39
|
40
|
36
|
36
|
39
|
42
|
46
|
51
|
50
|
51
|
51
|
50
|
|
76
|
66
|
54
|
60
|
62
|
61
|
|
TCL
|
35
|
37
|
39
|
34
|
35
|
35
|
38
|
44
|
48
|
49
|
48
|
48
|
46
|
76
|
|
63
|
53
|
58
|
60
|
59
|
|
Cdc42
|
40
|
40
|
42
|
37
|
37
|
38
|
43
|
43
|
53
|
51
|
50
|
53
|
56
|
66
|
63
|
|
61
|
69
|
71
|
70
|
|
RhoG
|
39
|
40
|
40
|
37
|
41
|
41
|
44
|
46
|
55
|
55
|
53
|
46
|
48
|
54
|
53
|
60
|
|
72
|
72
|
70
|
|
Rac2
|
41
|
42
|
40
|
39
|
40
|
39
|
46
|
50
|
53
|
53
|
54
|
50
|
54
|
60
|
58
|
69
|
72
|
|
92
|
89
|
|
Rac1
|
42
|
42
|
41
|
39
|
41
|
42
|
49
|
49
|
57
|
57
|
55
|
52
|
54
|
62
|
60
|
71
|
72
|
92
|
|
93
|
|
Rac3
|
41
|
41
|
40
|
38
|
39
|
40
|
49
|
47
|
55
|
54
|
54
|
53
|
54
|
61
|
59
|
70
|
70
|
89
|
93
|
|
Compared with other Ras superfamily proteins, the Rho-family small G proteins have a Rho insert region, which is a unique α-helical sequence located between the fifth β strand and the fourth α helix in the G domain
[5]. Many of the typical Rho-family GTPases only have a Rho-type G domain with short N- and C-terminal extensions
[6][7]. Similar to other Ras superfamily members, they also have a hypervariable region encoding a prenylation site at their C-termini and sometimes a polybasic region. These signals allow them to associate with specific membrane compartments
[8].
Additionally, some of them also possess a CAAX (C: Cys, A: aliphatic residue, X: any amino acid) tetrapeptide sequence at their C-termini, which is important for lipid modification. This process involves three main steps, which are isoprenylation, proteolysis and carboxyl methylation
[9]. An isoprenoid lipid is attached to the CAAX box by a prenyltransferase, such as geranylgeranyltransferase (GGTase) or farnesyltransferase (FTase). Both of these enzymes recognise the CAAX sequence before adding a 20-carbon geranylgeranyl or 15-carbon farnesyl to the cysteine residue of the CAAX sequence via a thioether linkage. Prenylation is followed by the proteolysis of the three C-terminal residues (AAX) by a prenyl protein peptidase (from the Rce1 family as an example) to release AAX. The prenylated cysteine is then methylated by isoprenyl-cysteine carboxyl methyl-transferase. Compared with the atypical small RhoGTPases, many of the typical Rho-family GTPases are geranyl-geranylated or farnesylated, and only a few, such as RhoB, TC10 and Rac1, are palmitoylated ()
[8][10]. However, some of the atypical small Rho GTPases also have a functional CAAX box that allows them to undergo prenylation (). For instance, both RhoU and RhoV only undergo palmitoylation, while Rnd proteins undergo farnesylation. Conversely, the RhoBTB subfamily does not possess any canonical CAAX motifs, suggesting that they may not undergo any lipid modification at the C-terminal.
Table 2. C-terminal sequences of the Rho-family small GTPases and their lipid modification.
|
Group
|
Rho Protein
|
C-Terminal Sequence
|
Lipid Modification
|
Ref
|
|
Typical
|
RhoA
|
KDGVREVFEMATRAALQARRGKKKSGCLVL
|
GG
|
[11]
|
|
RhoB
|
VREVFETATRAALQKRYGSQNGCINCCKVL
|
GG, F, P
|
|
RhoC
|
KEGVREVFEMATRAGLQVRKNKRRRGCPIL
|
GG
|
|
Rac1
|
RGLKTVFDEAIRAVLCPPPVKKRKRKCLLL
|
GG, P
|
|
Rac2
|
RGLKTVFDEAIRAVLCPQPTRQQKRACSLL
|
GG
|
|
Rac3
|
RGLKTVFDEAIRAVLCPPPVKKPGKKCTVF
|
GG
|
|
RhoG
|
QDGVKEVFAEAVRAVLNPTPIKRGRSCILL
|
GG
|
|
Cdc42
|
QKGLKNVFDEAILAALEPPEPKKSRRCVLL
|
GG
|
|
TCL
|
AVFDEAILTIFHPKKKKKRCSEGHSCCSII
|
F
|
|
TC10
|
DEAIIAILTPKKHTVKKRIGSRCINCCLIT
|
F, P
|
|
Atypical
|
RhoU
|
QQQPKKSKSRTPDKMKNLSKSWWKKYCCFV
|
P
|
[11]
|
|
RhoV
|
EHKARLEKKLNAKGVRTLSRCRWKKFFCFV
|
P
|
|
RhoD
|
AVFQEAAEVALSSRGRNFWRRITQGFCVVT
|
F, GG
|
[12]
|
|
RhoF
|
EDVFREAAKVALSALKKAQRQKKRRLCLLL
|
F, GG
|
|
Rnd1/ RhoS
|
LSKRLLHLPSRSELISSTFKKEKAKSCSIM
|
F
|
[11]
|
|
Rnd2/ RhoN
|
MQRSAQLSGRPDRGNEGEIHKDRAKSCNLM
|
F
|
|
Rnd3/ RhoE
|
KRISHMPSRPELSAVATDLRKDKAKSCTVM
|
F
|
|
RhoH/ TTF
|
VFECAVRTAVNQARRRNRRRLFSINECKIF
|
GG, F
|
[11][12]
|
|
RhoBTB1
|
KREREKEDIALNKHRSRRKWCFWNSSPAVA
|
Unknown
|
N/A
|
|
RhoBTB2
|
KRRWLFWNSPSSPSSSAASSSSPSSSSAVV
|
Unknown
|
Putative palmitoylated cysteines are in boldface type and coloured red and CAAX prenylation motifs are underlined. N/A: not applicable.
Among the Rho-family GTPases, the most extensively studied members are RhoA, Rac1 and Cdc42. These proteins and their subfamilies are known as classical or typical Rho-family GTPases. The other Rho-family proteins are known as the non-classical or atypical Rho GTPases and include the RhoBTB and Rnd subfamilies. This classification is made based on the ability of the Rho-family GTPases to undergo the standard GTPase cycle
[6][13]. The atypical Rho GTPases often contain extra domains and are therefore longer than classical Rho GTPases.
Compared with the atypical small GTPases are further divided into fast-cycling and GTPase-defective G proteins (). In resting cells, the typical Rho-family GTPases are generally assumed to be in their inactive GDP-bound form either alone or in complex with RhoGDIs
[14]. However, in contrast to the typical Rho-family GTPases, RhoD, RhoF, RhoU and RhoV have a rapid nucleotide exchange compared with standard GTP hydrolysis, which classifies them as fast-cycling small G proteins
[15][16][17]. The Phe28 mutations of Ras, Rac1, Cdc42 and RhoA have rapid nucleotide exchange and can induce oncogenic transformation
[18][19]. RhoU and RhoV also have mutated phenylalanine at position 28 (), and this factor might be one of the reasons for considering both of these proteins as fast-cycling small G proteins.
Table 3. Selected amino acid sequences of the typical and atypical small Rho GTPases.
|
Amino Acids 12, 59 and 61
|
|
Group
|
Subfamily
|
Member
|
Sequence
|
|
Classic
|
Cdc42
|
Cdc42
|
12 59 61
  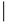
|
|
GDGAV---AGQED
|
|
Fast-cycling
|
RhoU/RhoV
|
RhoU
|
GDGAV---AGQED
|
|
RhoV
|
GDGAV---AGQDE
|
|
RhoD/RhoF
|
RhoD
|
GDGGC---AGQDD
|
|
RhoF
|
GDGGC---AGQED
|
|
GTPase defective
|
RhoBTB
|
RhoBTB−1
|
GDNAV---FGDHH
|
|
RhoBTB−2
|
GDNAV---FGDHH
|
|
Rnd
|
Rnd1
|
GDVQC---SGSPY
|
|
Rnd2
|
GDAEC---SGSSY
|
|
Rnd3
|
GDVQC---SGSPY
|
|
RhoH
|
GDSAV---AGNDA
|
|
Amino acids 28
|
|
Classic
|
Rac
|
Rac1
|
28
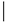
|
|
SYTTNAFPGEYIP
|
|
Fast-cycling
|
RhoU/RhoV
|
RhoU
|
SYTTNGYPTEYIP
|
|
RhoV
|
SYTCNGYPARYRP
|
|
RhoD/RhoF
|
RhoD
|
VFADGAFPESYTP
|
|
RhoF
|
VYSQGSFPEHYAP
|
Amino acid residues 12, 28, 59 and 61 are in boldface type and underlined [13].
The fast-cycling Rho GTPases have GTPase activity and are identical to Cdc42 at amino acids Gly12, Ala59 and Gln61 (Ras numbering) ()
[13]. Gly12 and Ala59 are important for GTP hydrolysis, while Gln61 is necessary to stabilise the transition state of hydrolysis
[20]. These amino acid residues are reported to be mutated in GTPase-deficient Ras
[21]. By contrast, the GTPase defective atypical Rho-family members, including Rnd1, Rnd2, Rnd3, RhoBTB1, RhoBTB2 and RhoH, do not possess the conserved Gly12, Ala59 and Gln61 compared with Cdc42 ()
[4][13].
Most studies have focused on the role of the typical small Rho GTPases in diseases, but information on the atypical small Rho GTPases is limited. Among the rare small Rho GTPases that receive much attention is RhoH as it is associated with several diseases, such as B cell malignancies, immunodeficiency and autoimmune diseases. This small GTPase has a unique role compared with others because it acts as a tumour-suppressor protein. Hence, a thorough understanding of the function of RhoH in both normal and disease conditions may help to identify and design an efficacious target.
2. RhoH, An Atypical Rho Family Small GTPase
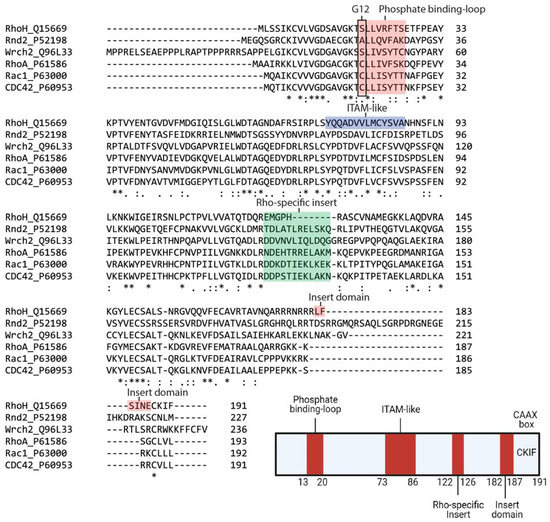
Within the family, RhoH and Cdc42 share 42% sequence similarity, with the major differences in the C-terminal region (). It also contains a shorter Rho insert region that is more similar to classical Rho-family proteins but is GTPase defective because conserved residues corresponding to Gly12, Ala59 and Gln61 (Ras numbering) are absent (). However, RhoH has a CAAX box, a CKIF motif at its C-terminus, which allows farnesylation and geranyl-geranylation
[22] and facilitates membrane targeting. RhoH myristoylation at the N-terminal may also assist RhoH attachment to the plasma membrane and promote its interacting partners, Lck, ZAP−70 and Syk recruitment to the membrane to facilitate T cell receptor (TCR) signalling
[23][24][25]. Among the Rho family members, RhoH is the only member with an ITAM-like motif, which is characterised by two tyrosines, spaced by six amino acids: Y73xxA76
6Y83xxA86 (). This consensus motif is crucial for the recruitment of ZAP−70 to CD3ζ in the immunological synapse
[23].
Figure 1. Sequence alignment and architecture of RhoH. Sequence alignment of RhoH, Rnd2, Wrch2, RhoA, Rac1 and Cdc42. Rnd2 and Wrch2 are representatives of the fast-cycling and GTPase-defective small GTPases, while RhoA, Rac1 and Cdc42 are typical small Rho GTPases (Top). RhoH shares a phosphate-binding loop with other small Rho GTPases but is assumed as GTPase defective due to the absence of Gly12. The Rho-specific insert in RhoH is shorter than that in other small Rho GTPases. RhoH also has an ITAM-like motif at position Tyr73 to Ala86 and a carboxyl-terminal insert domain (L182FSINE187) that function to regulate RhoH stability. RhoH also undergoes prenylation at the C-terminal due to the presence of the CAAX box (CKIF), similar to other typical small Rho GTPases (Bottom). (Created with Biorender).
Given that RhoH may be GTPase defective, other mechanisms have been described to regulate RhoH activity, including the regulation of mRNA level and tyrosine phosphorylation of the unique ITAM-like motif
[22]. Additionally, RhoH activity can be regulated by lysosomal degradation through its unique C-terminal region, L
182FSINE
187 domain, located in between its polybasic region and CAAX box. This insert domain has been shown to regulate RhoH stability via chaperone-mediated autophagy (CMA)
[26].
In contrast to the typical Rho-family members, which are best studied for their role in promoting actin cytoskeleton reorganisation, especially during cell division and migration
[2], RhoH does not regulate actin reorganisation in NIH3T3 or MDCK cells
[27]. However, Mino et al., (2018) showed that RhoH is involved in modulating the structure of actin-cytoskeleton and transcriptional activity during T cell migration and adhesion by forming a multi-protein complex with p120 catenin and the transcriptional regulator, Kaiso, to attenuate Rac1 signalling. Aside from regulating Rac1 signalling, RhoH interaction with Kaiso is necessary to facilitate Kaiso nuclear localisation to repress
BCL6 gene, a transcriptional repressor expression, resulting in an increase in tumour suppressor p53 protein
[28]. Collectively, the inhibitory function of RhoH would help to reduce the events of cell survival, migration and invasion
[29][30]. By contrast, although RhoH is assumed to be expressed only in haematopoietic cells, it was shown to promote cell migratory polarity in prostate cancer cell line by directing Rac1 and PAK2 to membrane protrusions
[31]. These contrary functions of RhoH might be dependent on the type and origin of the cells. For instance, RhoH is dispensable for the development of myeloid, erythroid, and B cells but is crucial for T cell production, survival and migration
[32][33]. This function is consistent with the study of Chae et al., (2010) that showed a lack of RhoH-impaired thymocyte development and stimulation of peripheral T cell unresponsiveness
[23][34].
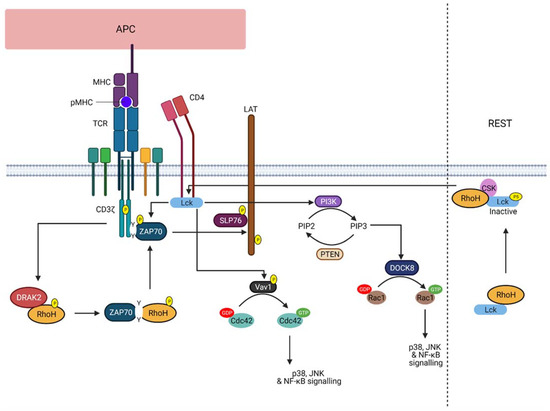
The current knowledge of RhoH function is only limited in T cells (), whereupon during response to TCR activation, RhoH is phosphorylated by kinases, such as DRAK2
[35]. This phosphorylation event promotes its interaction with ZAP−70 via the ITAM-like motif. ZAP−70 and Lck will then be recruited to the TCR, where ZAP−70 is activated and colocalised with its substrates. As a result, activated ZAP−70 will promote linker of activated T cells (LAT) and SLP76 signalling in T cell, crucial for T cell activation and development
[23][24][25][36]. Additionally, RhoH was also found to act as an adaptor protein that retains Lck in an inactive state, thereby suggesting that RhoH can regulate both pre-TCR and TCR signalling during T cell development. However, in response to ligand-mediated TCR activation, Lck is recruited to the membrane by RhoH and dephosphorylated by CD45, resulting in Lck auto-activation and its release from RhoH
[37]. Active Lck will then stimulate PI3K signalling and the activity of RhoGEF, Vav1. Nonetheless, despite the obvious role of RhoH in T cell development, RhoH involvement in T cell-specific malignancies, such as in T cell lymphomas, is not well understood. Conversely, RhoH is known to be associated with B-cell neoplasm, but its function in regulating B cell signalling should be further studied.
Figure 2. RhoH function upon TCR activation. The illustration shows the role of RhoH in regulating TCR signalling. Upon TCR stimulation, RhoH is phosphorylated by DRAK2 that promotes the interaction between RhoH and ZAP−70 via the ITAM-like motif of RhoH. ZAP−70 is recruited to the TCR CD3ζ chain, thereby activating LAT and SLP76 signalling. As for Lck, RhoH functions to assist Lck recruitment to the membrane to facilitate TCR signalling. Active Lck promotes PI3K signalling and PIP3-induced DOCK8 activation. As a result, Rac1-related signalling is activated. Vav1-induced Cdc42 activation can also be regulated by Lck. However, at rest, RhoH recruits Lck to the plasma membrane, where RhoH accelerates and supports the inhibitory phosphorylation of Lck at Y505 (labelled with P5) by CSK (Created with Biorender).
3. RhoH as a Therapeutic Target
Considering the importance of RhoH in regulating TCR and BCR signalling pathways, much effort has been exerted on targeting RhoH to control diseases derived from abnormal T and B cell activation, such as cancer, immunodeficiency disorders and autoimmune diseases. RhoH protein levels are frequently reduced in the aforementioned diseases, suggesting the need to rectify the aberrant RhoH expression in the haematopoietic cells. The current drugs known to modulate RhoH activities are lenalidomide and ibrutinib
[38]. However, the use of these drugs is critically dependent on the types of disease as these drug treatments were shown to regulate RhoH protein expression. For instance, the use of lenalidomide in the CLL murine model results in a decreased RhoH protein expression that promotes Rac1 and RhoA activation
[39]. Additionally, deregulated RhoH protein expression can be corrected by using the current approach that relies on the potential therapeutic opportunity to target CMA
[40]. This factor is based on the understanding that cellular protein levels are significantly influenced by protein stability. Hence, regulating their modes of degradation may promote protein stability. For instance, all-
trans retinoic acid can be used to block the inhibitory effect of retinoic acid receptor alpha (RARα) on the CMA process, especially on LAMP2A expression and its trafficking to lysosomes
[41]. This approach can be applied to RhoH as the presence of the inserted domain, LFSINE, at the C-terminal was shown to act as a recognition signal for lysosomal uptake and CMA-mediated degradation, while not affecting its function on both T cells and B cells
[26].
Another alternative is by targeting the RhoH interacting partner, ZAP−70. RhoH mediates TCR-induced ZAP−70 activation by recruiting it to CD3ζ in the immunological synapse
[23][32][34]. Additionally, RhoH was also found to promote ZAP−70-induced BCR signalling, which has been implicated in CLL
[33][42][43][44]. Hence, preventing ZAP−70 recruitment to the plasma membrane may help to block excessive signalling. Several studies have identified potential ZAP−70 inhibitors that can modulate the interaction with TCR
[45]. However, despite its therapeutic potential in autoimmune diseases and organ transplant rejection in vitro and in vivo, these inhibitors have never been tested in clinical trials, indicating the need for further investigation.
Alternatively, a peptide that can block or stimulate RhoH-related signalling may be used as a potential therapeutic target. The use of therapeutic peptides is an emerging targeted therapy that is often associated with rapid production and capacity for modification. However, this strategy has several drawbacks that usually arise due to their limited bioavailability and specificity
[46][47]. Thus, far, no specific RhoH-therapeutic peptide is available or has been reported that may be due to the limited knowledge of RhoH functional activities.