1000/1000
Hot
Most Recent

Cisd2 (CDGSH Iron Sulfur Domain 2) is an oxidative stress-sensitive gene, the expression of which is able to prolong the lifespan in mice.
Cisd2 (CDGSH Iron Sulfur Domain 2) is an oxidative stress-sensitive gene, the expression of which is able to prolong the lifespan in mice [1]. Cisd2 loss-of-function mice exhibit premature aging phenotypes and have a shortened lifespan. Conversely, Cisd2 transgenic mice not only are longer lived (both males and females), they also have a healthier physical condition, such as better fur function, increased muscle strength and improved cardiac function [2][3]. The Cisd2 protein has been localized to mitochondria, mitochondria-associated membrane (MAM) and the endoplasmic reticulum (ER), and is involved in calcium homeostasis [4]. We have demonstrated using aged mice that maintenance of the expression level of Cisd2 sustains metabolic activity, ameliorates aging-associated mitochondrial dysregulation, reduces DNA damages and improves the calcium imbalance within skeletal muscles [2], liver [4] and heart [3]. Taken together, Cisd2 is a lifespan regulator and its expression level seems to be a critical factor in relation to prolong healthspan.
Cisd2 and exercise have both been reported to contribute to an extended lifespan and to improve healthspan. A recent report has shown that the protein levels of Cisd1 and Cisd2 in skeletal muscle and white adipose tissue are increased approximately 1.5-fold and 1.2-fold after 4 weeks of voluntary excise in mice [5]. The authors also observed that there were significant increases in the levels of multiple mitochondrial proteins, which agrees with our previous discovery of increased mitochondrial number in the muscle of Cisd2 transgenic mice compared to their wild type cohorts [2]. To examine the transcription of Cisd2 in real-time, here we have generated a Cisd2 reporter transgenic mouse that carries luciferase as the reporter, which is under the control of the mouse Cisd2 endogenous upstream 31.3-kb promoter sequence (i.e., a Cisd2-Luc mouse). In addition to the endogenous promoter, a 46.9-kb region downstream of Cisd2 coding region was also included in the transgene construct in order to preserve the regulatory elements of the Cisd2 gene. The Cisd2-Luc reporter mice were trained on a treadmill for 56 days. It was found that a drastic enhancement in Cisd2 transcription was able to be observed. The most intense signal was observed at the abdomen, with the thymus showing the next largest increase in signal. A moderately increase in signal was also observed in forelimbs and hindlimbs (Figure 2A).
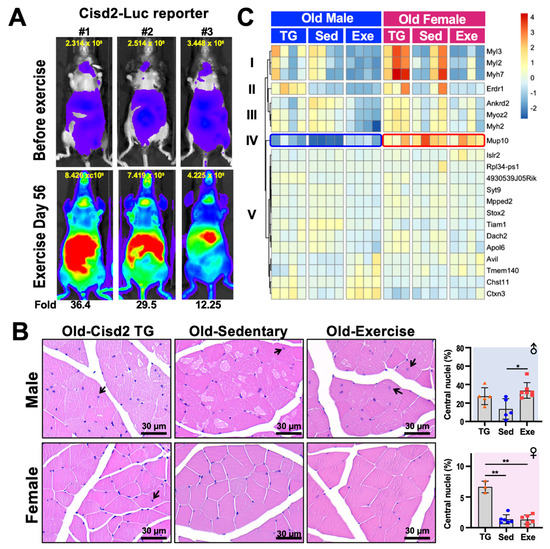
Figure 2. Histology and transcription analysis of femoris muscle from mice that have practiced long-term exercise. (A) Luciferase activity in Cisd2-Luc reporter mice before and after 56-days of treadmill exercise. The treadmill protocol was 6 m/min for 5 min, 9 m/min for 5 min, 12 m/min for 20 min, 15 m/day for 5 min and 12 m/day for 5 min, for a total of 450 m/day. The Cisd2-Luc reporter mice ran on treadmills at a frequency of 5 days/week for 8 weeks (56 days). The quantification of luciferase intensity in each of the mice in the figure is shown at top of each image. Increased (fold change) Cisd2-Luc luciferase signals are numbered below the images; these represent mice after the exercise was completed. The photos were taken ventrally. (B) H&E staining of the femoris muscles of Cisd2-TG, Old-Sed and Old-Exe male and female mice. The arrows indicate myofibers that contain central nuclei. Bars: 30 μm. Percentage of central nucleation in each group of mice is presented on the right. The data are presented as means ± SD. Statistical analysis used the Student’s t-test. *, p < 0.05; **, p < 0.01. TG, Cisd2-TG; Sed, sedentary; Exe, exercise. (C) A heatmap showed the differentially expressed gene (RNA-Seq) clusters for the male and female femoris muscle that have been changed by long-term treadmill exercise. The RNA-seq data was analyzed by “DEseq2” and the DE genes were defined as LFC > 0.14 and FDR < 0.1. The heatmap was generated using the “pheatmap” R package (Raivo Kolde (2019). pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap) according to rlog transformed count calculated by “DEseq2”.
We further analyzed the histology of the femoris muscle of male and female Old-Sed and Old-Exe mice as described in Figure 1, and compare these to the Cisd2-transgenic (Cisd2-TG) mice at 24–26 months old as reported in Shen et al. [4]. Quadriceps femoris is composed of four individual skeletal muscles, rectus femoris, vastus medialis, vastus lateralis and vastus intermedius. The quadriceps femoris was chosen for this study because of its mass, the fiber type composition and our recent use in other studies. The quadriceps femoris is the largest muscle present in the mouse hindlimb by mass [6]. Furthermore, the quadriceps femoris muscle in terms of fiber types represent the majority of skeletal muscle. The fiber type of the quadriceps femoris muscle is IIx/IIb in rodents and IA/IIA and IIA/IIX in humans, which are commonly referred to as fast-twitch fibers. Most other skeletal muscle in limbs, other than the soleus muscle, have a similar fiber type composition both in humans and other mammals [7]. Finally, the quadricep femoris has often been used to study exercise [7] and the effects of aging [4][8][9] in mouse and humans. In agreement with the results on systemic metabolism (Figure 1B), we found a gender difference in the histology of skeletal muscle, particularly in the Old-Sed mice. In the male mice, multiple vacuolated fibers were observed in the skeletal muscles of these old sedentary mice. This pathology was obviously attenuated in old Cisd2-TG male mice [3] and the Old-Exe male mice (Figure 2B). Additionally, central nucleation was seen in the femoris muscle of Cisd2-TG (27.3 ± 9.15 % of total fiber, p = 0.07) and Old-Exe (33.6 ± 8.50 % of total fiber, p = 0.01) male mice (Figure 2B, arrows). Central nucleation in muscle cells is an indicator of degeneration followed by regeneration. Accordingly, these findings suggest that exercise and an elevated level of Cisd2 are both able to enhance the repair of muscle fibers. However, in female mice, the age-related pathohistological alterations to the skeletal muscles is less obvious in the Old-Sed group and thus there appears to be no differences in pathology across the three groups of female mice (Figure 2B). Notably, quantification of central nucleation in the female mice reveals that Cisd2-TG mice exhibited a significantly higher ratio of central nuclei in their muscle fibers (6.63 ± 1.0 % of total fiber) (Figure 2B). This suggests that Cisd2 indeed has a beneficial effect on muscle regeneration within both males and females during aging.
To better understand the alternations in the transcriptome that are brought about by long-term exercise and/or Cisd2 overexpression, as well as which signaling pathways are potentially involved during muscle aging, we performed RNA-seq analyses by DEseq2 [10] on femoris muscles obtained from the various groups of mice, namely Old-Exe, Old-Sed and Cisd2-TG (24–26 months old) mice. Due to the substantial individual variation often observed in tissue samples collected from aged mice, a less stringent criteria [log2 fold change (LFC) > 0.14 and false discovery rate (FRD) < 0.1] were set in order to identify the differentially expressed (DE) genes. Comparing Old-Exe and Old-Sed femoris muscle, 19 DE genes and two DE genes were identified in males and females, respectively. On the other hand, only one DE gene, namely 4930539J05Rik, was found when Cisd2-TG and Old-Sed mice were compared among males, and no DE gene was identified among females. These findings correlate with the macroscopic observations obtained by histopathological analysis (Figure 2B).
A heatmap of the DE genes was plotted to explore the expression pattern of these genes across the three groups (i.e., Cisd2-TG, Old-Sed and Old-Exe, Figure 2C). There were 21 DE genes in total and these were able to be grouped into five clusters. Cluster I, II and III consisted of the DE genes that are significantly down-regulated in the Old-Exe group, compared to the Old-Sed group, and most of those DE genes encode sarcomere proteins. Of these, Myl3 (LFC = −5.48, FDR = 0.01), Myl2 (LFC = −4.3, FDR = 0.06) and Myh7 (LFC = −4.94, FDR = 0.09), encode the slow-twitch type myosin, and Myh2 (LFC = −2.61, FDR = 0.001), encodes fast-twitch type of myosin [11][12][13]; Myoz2 is a gene mediating fiber type switching, and Myoz2 deficiency has been reported to cause an increase in slow-twitch muscle fibers [14]; Ankrd2 encodes a I-band structure protein that also participates in embryonic myoblast differentiation [15][16][17].
The expression profile of Mup10, the only gene in cluster IV, is particularly interesting. Compared to male Old-Sed mice, Mup10 is significantly up-regulated in Old-Exe (LFC = 3.54, FDR = 0.008) and Cisd2-TG (LFC = 2.98, FDR = 0.201) male femoris muscle (Figure 2C, blue frame). Mups (major urinary proteins) form a highly polymorphic gene family that are expressed in the liver. They are secreted into the bloodstream in order to regulate pheromone stability, and urine Mup profiles are able to function as individual identity signatures of the owners. Circulating MUPs serve as a metabolic signal that links the regulation of glucose and lipid metabolism [18]. Whereas the protein function of Mup10 has not been extensively explored, recombinant MUP1 has been shown to elevate energy expenditure markedly, ameliorate hyperglycemia and reduce glucose intolerance in mice with type 2 diabetes [18]. Intriguingly, the level of Mup10 transcripts in females is higher overall than in males (Figure 2C, red frame), whereas many other reported MUPs show much higher transcription levels in males than in females [18]. These findings suggest that gender dimorphism of Mup10 in rodents is significantly associated with exercise.
Differences of the 13 DE genes in cluster V were moderate between the three experimental groups. Among these genes, Islr2 (LFC = 2.05, FDR = 0.005) and Rpl34-ps1 (LFC = −22.09, FDR < 0.001) are the DE genes in the female Old-Exe group compared to the gender-matched Old-Sed group, while 4930539J05Rik (LFC = 2.39, FDR < 0.001) is the only gene differentially expressed in male Cisd2 TG muscle compared to Old-Sed muscle. In addition, Rpl34-ps1 can be seen to have a gender difference in its expression profile across exercised mice; this gene is significantly up-regulated in male Old-Exe (LFC = 15.8, FDR < 0.001) but down-regulated in female Old-Exe (LFC = −22.09, FDR < 0.001). Finally, Avil (LFC = 1.48, FDR = 0.06), Tmem40 (LFC = 1.34, FDR = 0.07), Chst11 (LFC = 1.08, FDR = 0.07) and Ctxn3 (LFC = 1.66, FDR < 0.001) are all up-regulated in Old-Exe femoris muscle samples compared to the Old-Sed group. Of the last group of genes, it is known that Chst11 (carbohydrate sulfotransferase), a Wnt/β-cateinin down-stream target [19], participates in chondrocyte development and cartilage morphogenesis [20]. Collectively, our findings reveal that at middle-to-old age treadmill exercise creates a clear gender dimorphism in the transcriptome of the femoris muscle, and that this specifically bring about and causes expression changes in fast-twitch myosins, slow-twitch myosins and the secreted/excreted protein Mup10.