1000/1000
Hot
Most Recent

Transcription of mRNA is carried out by RNA ´polymerase II in eukaryotes. This a complex process involving protein biopolymers. mRNA can be used as vaccine and can be delivered into the cells to produce an antigen. This mRNA-based vaccine could be used for several infectious diseases, cancer and SARS-CoV-2 virus. It can be also used to deliver a missing protein in a particular cell type. A functional mRNA can be in vitro transcribed, either by using a viral RNA polymerase or an eukaryotic RNA polymerase II together with a capping system.
Essentially, transcription of eukaryotic mRNA is the process of copying an RNA, using it as a template of the DNA, which is carried out by the enzymatic protein biopolymer RNA polymerase II (RNAPII). The template DNA is a double helix and the RNAPII transcription machinery must recognize the promoter element on the coding strand (or the mRNA-like strand) and copies the mRNA on the template strand (Figure 1). The transcription process is mainly carried out in three steps, which are initiation, elongation, and termination [1].
The conventional vaccination is one of the major breakthrough in modern Medicine, which have reduced the incidence of infectious diseases such as measles and completely eradicating others such as smallpox. However, conventional vaccination has not been effective against another infectious diseases such as influenza or other emerging diseases such as those caused by Zika and Ebola viruses. Nowadays, we are facing a devasting pandemic all around the world caused by the SARS-CoV-2 virus which causes the COVID-19 disease. This pandemic has caused millions of infected people and several thousands of deaths. Additionally, this pandemic is devasting the economies of most of the countries. mRNA-based vaccines could have a great impact to fight this virus, since mRNA vaccines are faster and cheaper to produce than conventional vaccines. In addition, this process can be standardized and scaled to allow a quick response to this pandemic. Besides, an mRNA-based vaccine is safer both for the personal that produces it and for the patient, since it does not use highly infectious viruses. Taken altogether, we suggest a strategy (Figure 2) to develop an mRNA-based vaccine against SARS-CoV-2 by targeting the spike S protein (S protein) which binds to membrane ACE2 (angiotensin-converting enzyme 2) receptors. ACE2 receptors can be found in several organs such as lungs, heart, kidneys and the gastrointestinal tract, which are the SARS-CoV-2 targets. We could expect that once the mRNA encoding the S protein is delivered into the myocytes of the muscle, it will be translated and secreted and taken up by macrophages, dendritic cells and other cells of the immune system to be presented to T and B cells. Once a B cell response is mounted, the resulting antibodies could block the binding of the virus to the ACE2 receptors and thus prevent the virus to enter to the target cells. In addition, activated T cytotoxic cells can help to destroy the invading virus (Figure 2).
Figure 2. Schematic procedure to produce an mRNA vaccine against SARS-CoV-2. From the analysis of the amino acid sequence of the S protein an antigenic region can be predicted. Whenever possible, a region of the S protein which is less prone to mutations should be selected. The amino acid sequence of the selected region can be back translated to DNA nucleotide sequence and it can be chemically synthetized according to the codon usage. Afterwards, this DNA sequence can be cloned into a DNA plasmid vector, as described in Figure 3, and used for in vitro transcription (IVT) by using a RNA polymerase to produce an mRNA that can be 5′ capped to be fully functional. This mRNA can be coated with lipid nanoparticles and delivered to the myocytes to be translated by the cell ribosomes. The resulting antigens will be secreted and take up by macrophages and dendritic cells, which will process and present the antigen to the immune system cells to activate both humoral (antibodies) and cellular (cytotoxic) immune responses.
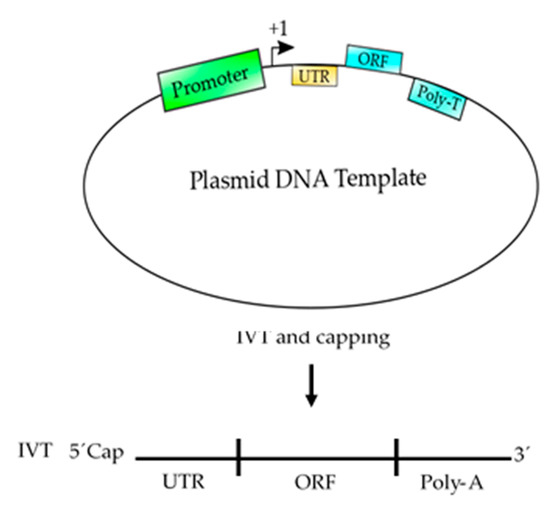
Figure 3. Schematic representation of a plasmid DNA template used to produce an mRNA-based vaccine. This template contains the basic elements to produce a functional mRNA. A promoter is needed for the RNA polymerase to start transcription at the +1 site. The 5′ UTR is added to enhance the translation and the stability of the mRNA and the ORF contains the coding sequence of the antigen, which could be synthetized according to the codon usage to enhance the translation process. Finally, a poly T of at least 200 nucleotides is needed to produce a poly A tail on the mRNA. After, the template is transcribed (in vitro transcription, IVT) using RNA polymerase and ribonucleotides to produce an mRNA, which should be 5′ capped to produce a functional mRNA to be used as vaccine.
Messenger RNA can be used in Biotechnology and Medicine as RNA vaccines against infectious diseases and also in cancer immunotherapy to deliver multiple antigens with one immunization [2][3]. RNA vaccines might have several advantages over DNA-based vaccines, since they only need to reach the cytoplasm, are more immunogenic and do not possesses oncogenic potential via integration into the host genome [2][3].
Typically, RNA-based vaccine consists in a functional mRNA encoding an antigen, which is delivered into the cellular cytoplasm of target cells where it can be translated via poly-some formation [4]. There are different non-viral strategies for the delivery of mRNA-based vaccines, including naked mRNA vaccines, gene gun delivery method, protamine condensation of the mRNA, adjuvants and encapsulated mRNA-based vaccines. The strategy of using naked mRNA vaccines uses only mRNA formulated in a buffer and injected directly into the individual [5]. The gene gun delivery is an alternative method of mRNA delivery directly into the cell cytoplasm [6]. Specifically, it is a nanotechnology method in which the mRNA is coated onto gold particles, which are then accelerated toward a stopping plate by a helium pulse and penetrated into the cytoplasm of target cells [6]. Another method to improve mRNA stability is incubating it with protamine and inject this protamine-protected mRNA into the individual [7]. Additionally, the mRNA can be injected with molecules such us poly I:C RNA and CpG- containing motif molecules [8]. Lastly, another method, based on nanotechnology, has been used to deliver mRNA-based vaccines. Nanoparticles of cationic liposomes have been used to protect and deliver mRNA vaccines into the individuals [9][10].
The mRNA-based vaccines can be used to immunize against infectious diseases and several types of cancer [11]. Immunization against the Zika virus and against the rabies virus with mRNA-based vaccines are only two cases where this immunization procedure shows promising results [11][12][13]. Both mRNA-based vaccines were delivered by using nanoparticles of cationic liposomes in mice. The vaccines provided protection against lethal doses of virus in those immunized mice [11][12][13]. On the other hand, mRNA-based vaccines have been used to immunize against several types of cancer [2][3]. This strategy relies on the generation of a host immune response against specific tumor-associated antigens by the cytotoxic T cells, which are able to recognize and kill tumor cells. Once the mRNA-based vaccine is delivered into the cytoplasm of antigen presenting cells (mainly macrophages), the resulting antigen is presented together with MHC class I molecules to CD4+ T cells, activating a cellular response that leads to the destruction of the tumor cells. Additionally, the mRNA can act as an adjuvant and provide costimulatory signals via toll-like receptors. To illustrate this strategy, we describe two examples. An mRNA-based vaccine against malignant melanoma was developed using the injection of naked mRNA encoding melanoma-associated antigen [14]. In addition, another mRNA-based vaccine was developed against triple-negative breast cancer by delivering mRNAs encoding tumor-associated antigens into cationic liposomes [15].
Usually, the mRNA is synthetized “in vitro” using bacteriophage RNA polymerases, which are highly actives. However, these RNA polymerases can only synthetize an mRNA up to 5 Kb long. Eukaryotic RNAPII is able to synthetize mRNAs over 50 Kb or longer. In addition, RNAPII is less error prone than bacteriophage RNA polymerases, which is important to avoid mutations or the creation of stop codons.
By the present, all TFIIs necessary for transcription are cloned and expressed and it is possible to purify core RNAPII, thus allowing us synthesize mRNA in a test tube with all proteins of eukaryotic origin. Recently, Fujiwara and Murakami have reported a method to assemble large amounts of PIC using purified TFIIs and RNAPII from budding yeast [16]. Those PICs were assembled and purified further using glycerol gradient sedimentation [16]. We have also been attempting to assemble PICs from fission yeast by using whole cell extracts, which contain a full set of TFIIs, RNAPII and regulatory factors (Maldonado, E., unpublished results). For those experiments we used a DNA template containing a super core promoter (SCP) based on the work of Gershon and collaborators [17] and we added a GAL4 binding site upstream of the TATA box to allow strong in vitro transcription activation by the powerful GAL4-VP16 chimeric transcription factor. Those PICs have been assembled and further purified by the use of glycerol gradient sedimentation. The isolated PICs are highly active in transcription initiation and elongation and RNAs of more than 2 Kb long can be obtained. We think that by the use of similar methods it is possible to obtain large amounts of in vitro transcribed mRNA using eukaryotic RNAPII. Moreover, eukaryotic methyltransferase and capping enzymes can be added to the in vitro reaction together with precursors for the m7G. This point is very important since it has demonstrated that capping is a co-transcriptional process [18][19].
In the near future, we could image that a multitalented recombinant polypeptide of eukaryotic origin would be able to recognize the promoter in the DNA template and perform the elongation of the mRNA chain. To engineer this multitalented polypeptide, we must know all fundamental and necessary domains in each of the individual subunits of core RNAPII and TFIIs involved in the recognition of the promoter and in the function of transcription elongation. Moreover, another polypeptide containing methyltransferase and capping activities could be engineered to add the m7G cap on all the mRNAs to produce a template 100% efficient for translation. This would be an important advance on protein biopolymers able to synthesize mRNA for uses in mRNA-based vaccines.
In this review, we have described the PIC formation process on eukaryotic TATA-containing promoters and additionally the PIC complex formation on two eukaryotic non-TATA promoters, such as HomolD box and INR-containing promoters, in which the mechanisms of PIC formation are not fully understood yet.
A strategy is outlined to develop an mRNA-based vaccine to fight the SARS-CoV-2 virus, which is causing the devasting COVID-19 disease at the present-days. In addition, we reviewed the main applications of the mRNA-based nanomedicines and their potential therapeutic uses. Some of the mRNA-based vaccines have moved forward to clinical trials. In the case any mRNA-based vaccines were successful; these processes will be streamlined to establish a large scale-production platform and in near future mRNA vaccines will be used in humans and animals as well.
The main problems facing in vitro transcription of mRNA is the 5′ capping of the transcribed mRNA and the size of the transcript that can synthetize a viral RNAP. We have suggested an in vitro system using eukaryotic RNAPII, which is able to synthetize longer transcripts of up to 50 Kb long. The in vitro transcription setup is highly active and includes the capping system, which can synthetize the m7G cap in a co-transcriptional manner and makes the whole process more effective. Since, the in vitro transcription system used is rather simple, the functional mRNA product can be easily purified and used as a vaccine.