1000/1000
Hot
Most Recent

Colorectal cancer (CRC) is the third most frequent cancer in men and the second in women. The prognosis depends not only on the stage at diagnosis, but also the surgical alternatives and the systemic treatment received. Due to the implementation of screening programs, the introduction of novel systemic therapies, and the advanced surgical procedures, the oncological outcomes have dramatically improved in the last years.
Colorectal cancer (CRC) is the third most frequent cancer in men and the second in women. The prognosis depends not only on the stage at diagnosis, but also the surgical alternatives and the systemic treatment received. Due to the implementation of screening programs, the introduction of novel systemic therapies, and the advanced surgical procedures, the oncological outcomes have dramatically improved in the last years. However, CRC is still the second leading cause of cancer-related mortality worldwide [1].
In patients with localized CC, the assessment of risk of recurrence following surgery is a crucial point to indicate systemic adjuvant therapy. This assessment is based on tumor TNM staging and other clinicopathological characteristics including CEA status or lymphovascular and perineural invasion [2]. Currently, standard fluoropyrimidine-based therapies estimate an increase in overall survival of approximately 5% in stage II patients and 20% in stage However, at least 50% of patients with stages
Treatment paradigm is slightly different in locally advanced rectal cancer (LARC). Patients with clinical stage T3/4 or node-positive tumors are treated with neoadjuvant chemo-radiotherapy (CRT) or total neoadjuvant treatment (TNT) [3][4] followed by surgery. For those patients who received neoadjuvant CRT, the role of adjuvant chemotherapy is controversial, and FOLFOX only demonstrated benefit in stage
Moreover, it is essential to consider that these treatments are not exempt from adverse effects, such as digestive toxicity or palmoplantar erythrodysesthesia which can be fatal in a small subset of patients carrying a DPYD polymorphism [5], or peripheral neuropathy caused by oxaliplatin that limits day-to-day activity and is permanent in at least 10% of patients [6][7].
Thus, determining the presence or absence of minimal residual disease (MRD) is of utmost importance to guide clinicians to avoid both under- and overtreatment in the adjuvant setting. Circulating cell-free DNA (cfDNA) is a highly fragmented DNA mainly derived from apoptotic cells, predominantly apoptotic leukocytes, found in the blood. concentrations in healthy individuals range between 1 and 10 ng mL−1in plasma [8][9]. Circulating tumor DNA (ctDNA) is a fraction of cfDNA characterized by the presence of tumor-specific genomic alterations with a half-life of a few hours allowing for a real-time, non-invasive characterization of the tumor molecular profile.
ctDNA allows for a better and less aggressive characterization of the spatial tumor molecular heterogeneity and its temporal evolution compared to the traditional use of tissue tumor biopsy. ctDNA, although generally obtained from peripheral blood, can also be isolated from other fluids such as urine, saliva, or cerebrospinal fluid [10][11][12][13][14]. In the metastatic setting, a high concordance between detection of mutations in tissue tumor compared to ctDNA has been reported [15][16][17]. However, ctDNA represents between 0.005% and 11.7% of the whole cfDNA shed in the bloodstream [18] depending on the tumor size, tumor growth rate, and cell turnover [19].
Recently, there have been several advances in the development of new technologies for the detection of ctDNA. Current techniques allow for the detection of genomic molecular alterations (including point mutations, short insertions and deletions, copy number alterations, and fusions), as well as epigenomic changes (i.e., methylation) and cfDNA fragmentation pattern identification. The increased sensitivity in the detection of these biomarkers is crucial for an accurate detection of MRD in CRC cancer patients.
Several ongoing clinical trials are evaluating the use of ctDNA to guide adjuvant therapeutic strategies in CRC patients after curative-intent surgery, and will ultimately shed light onto whether ctDNA is a useful tool to improve outcomes of localized CRC patients. This review highlights and explores the current situation on this topic and its potential application in routine clinical practice.
The molecular landscape of colorectal cancer has been well characterized in the past decades, including chromosomal aberrations such as copy numbers alterations (CNAs), inversions, translocations, insertions, and deletions, as well as single nucleotide point mutations [20]. Epigenomics, referring to covalent modifications of DNA that result in a change in its function or in the regulation of the affected genes without altering the primary sequence, has also been well described in colorectal cancer. These molecular alterations are highly specific to cancer, and, thus, their detection in an individual’s blood potentially indicates the presence of cancer. especially in early stages of the disease makes its detection challenging [21][22][23].
Currently, two main strategies are used to study tumor genomic material in ctDNA for MRD detection after curative-intent surgery. On the one hand, techniques based on the detection of one or several mutations previously found in the primary tissue tumor. (mutation needs to be known ahead of time) and that only a limited number of known mutations can be tracked in the blood (although multiplexing assay is possible). However, it is a very sensitive (0.01%) and specific technique in addition to being rapid and cost-effective [24][25][26].
The second strategy, the so-called deep next-generation sequencing (NGS), allows for the detection of multiple genetic alterations in one sample. NGS conducts a non-directed scan by analyzing the entire genome to detect CNAs or point mutations through whole genome sequencing (WGS) or exome sequencing (WES) [27]. NGS has a high false discovery rate that requires pre-sequencing barcoding and post-sequencing bioinformatics for error suppression. In order to obtain a high sensitivity (0.1%), it is necessary to perform DNA barcodes or UMIs (unique molecular identifiers), initial DNA molecules with 10–12 random bases, such that the barcode is amplified and sequenced together with the DNA.
Healthy individuals, with age, acquire new mutations in hematopoietic cells [28] and, when lysed, can release cfDNA [29] sought to develop a protocol for the distinction between mutations of tumor origin and those of hematological origin. The authors found that half of the mutations identified in cfDNA of cancer patients were originated in the hematological compartment as a result of clonal hematopoiesis and were therefore not derived from the primary tumor. This study shows the need to sequence the leukocyte fraction in order to discriminate the origin of the mutation detected in cfDNA.
Currently, new strategies are being studied to increase the sensitivity of diagnostic techniques. These include the analysis of methylation tumor profile via nucleosomal positioning or epigenomic alterations at transcription factor binding sites [30][31]. Methylation patterns that inhibit gene expression are related to the tissue to which they originally belong and therefore reflect the origin of circulating DNA. using two methylation markers (WIFI and NPY) by ddPCR in CRC patients [32][33].
New NGS strategies are arising by combining different methods to improve ctDNA detection. Such is the case of LUNAR-1 technology (Guardant RevealTMGuardant Health), which integrates assessment of somatic alterations with an epigenomic cancer signature without a priori knowledge of tumor mutation. Sequencing data files are analyzed using a proprietary bioinformatics pipeline software to exclude common sources of interference such as CH of indeterminate potential [34]. However, the need of bioinformatics analysis to exclude eventual false negative results increases the complexity and subsequently its costs and turnaround times.
Regarding fragmentation, it is well described that the fragmentation cut-off points of circulating DNA are not random and are different for ctDNA compared to cfDNA from normal cells. While the mechanisms of this different fragmentation pattern are yet unknown, they help to characterize whether cfDNA is of tumoral origin or not (Figure 1).
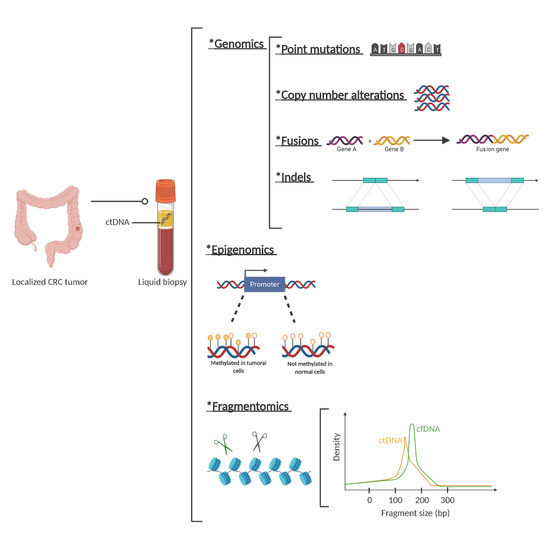
Figure 1. Circulating tumor (ct)DNA features. Description of genetic, epigenetic, and fragmentomic alterations that can be found in plasma cell free DNA analysis. *: The asterisks are placed as a way of listing the different techniques.
Emerging data suggest that elucidating nucleosome positioning opens promising new perspectives to identify the tissue source of origin of cancer from cfDNA, with an important clinical value to classify cancers and, to a further extent, to characterize, for example, cancers of unknown origin [35].
Single biomarkers in liquid biopsy often do not accurately predict disease status due to heterogeneity between individuals. To address this challenge, investigators are combining multiplexed measurements of different biomarkers that together define robust signatures for specific disease states. Machine learning approaches have been proposed to classify the cell of origin based on somatic mutation profiles in the genome of solid tissue biopsies. Therefore, it is crucial to investigate the applicability of sparse somatic mutation profiles in the identification of ‘cell of origin’ and explore potential improvements of the data analysis and prediction models to overcome sparsity [36].
With only few years, liquid biopsy has rapidly evolved from detecting point mutations in known given genes, to large genome sequencing and the detection of methylations and fragmentomics. This dramatic change has allowed us to improve ctDNA detection sensitivity and even avoid tissue information and carry out molecular studies only on liquid biopsy. Despite these technical improvements, liquid biopsies still have some biological limitations. In the metastatic CRC setting, the discordance in the finding of mutations between tissue and plasma has been shown to be related to the location of the metastasis (pulmonary or peritoneal) and the histology of the tumor (predominantly mucinous) [16][37][38].
after curative-intent surgery in patients with localized disease has been very consistently associated with a high risk of recurrence in different tumor types [39][40][41][42]. Several observational studies have shown that ctDNA detects MRD and is associated with recurrence in patients with localized CRC that have undergone surgery of the primary tumor. In addition, ctDNA has been shown to also be a useful tool for patients with metastatic CRC receiving multi-therapy with curative intention [18]. after surgery have a much lower probability of recurrence and eventual relapse tends to occur later in time.
Up to 25% of newly diagnosed CRC cases are stage II. While TNM staging remains the most relevant criteria for risk assessment after surgery, in stage II CC other clinicopathological parameters need consideration to decide the need for adjuvant chemotherapy [2]. Minor prognostic parameters for stage Identification of an accurate prognostic biomarker would help for a better selection of patients that benefit from adjuvant systemic therapy, and spare chemotherapy in cured patients.
With the aim to use ctDNA as a prognostic biomarker, Tie et al. [43] studied the impact of post-operative ctDNA detection in stage II colon cancer patients.
A total of 250 patients with stage II CC were included, in which a blood sample with ctDNA and CEA analysis at 4–10 weeks post-surgery (post-IQ) and subsequently every 3 months for 2 years was performed. Patients were given chemotherapy at the investigator’s criteria and a clinical follow-up was performed every 3 months including CT imaging tests every 6 months for 2 years. During the follow-up, 34 patients out of 230 (14.8%) had radiological recurrence, including 27 of 178 (15%) patients not treated with chemotherapy and 7 of 52 (13%) patients treated with adjuvant chemotherapy.
Seventy-eight percent (11 out of 14 patients) of patients who did not receive adjuvant treatment and had post-surgery positive ctDNA had radiological recurrence, whereas only 16 out of 164 (9.8%) ctDNA negative patients recurred. CEA was elevated in 7.4% of recurrent patients (2 of 27 cases) and none of the patients with positive ctDNA had an increase in CEA after surgery. Recurrence-free survival (RFS) was estimated to be 0% at 3 years in patients with post-surgery positive ctDNA, compared to 90% in patients with post-surgery negative ctDNA
The use of ctDNA was shown to be superior to the currently used clinicopathological risk parameters to decide on adjuvant treatment. On the other hand, the probability of recurrence was very low for patients with ctDNA negative, defining a subset of patients that could avoid adjuvant chemotherapy and potential associated toxicities.
Combining the use of ctDNA with imaging tests in the follow-up of these patients may help in the early detection of disease recurrence and potentially ultimately lead to better results in terms of survival [44][45].
Randomized phase III clinical trials have shown that the use of adjuvant chemotherapy with oxaliplatin plus a fluoropyrimidine (FOLFOX or XELOX regimens) improves the overall survival of patients with stage III CC [46]. Most of these patients receive chemotherapy, although up to 50% of them are cured by surgery, and chemotherapy could be potentially be spared [47][48]. Among patients receiving chemotherapy, approximately 30% will have recurrence of the disease and are potential candidates for other systemic treatments [49][50].
Retrospective subgroup analysis of large randomized clinical trials supported a different duration of adjuvant FOLFOX/ XELOX treatment depending on pathological risk factors. Thus, in patients considered to be at low risk ( the duration of the adjuvant treatment of 3 months is considered appropriate, whereas in the case of patients with high-risk tumors (T4 and/or N2), the duration of treatment is recommended to be extended to 6 months [51].
as a biomarker in patients with stage I to III CRC (mostly stage III) with the aim of demonstrating that the presence of post-surgery ctDNA is related to a high probability of recurrence. The study enrolled 130 patients and plasma samples were collected before surgery, after 30 days, and then every 3 months up to 3 years. The recurrence rate was 70% (7 out of 10 patients) in patients with positive ctDNA after surgery, compared to 11.9% (10 patients out of 84) for patients with no ctDNA detection. After surgery, patients with ctDNA were 7 times more likely to relapse than ctDNA-negative patients (HR 7.2; 95% CI 2.7–19.0;p
In line with the aforementioned, the Australian team led by Tie [52] has published the results of a multicentric clinical trial in which 100 patients with a diagnosis of CRC stage III with the provision of administering adjuvant treatment with chemotherapy for 6 months were consecutively recruited. Samples were collected from the primary tumor and later peripheral blood samples for ctDNA determination 4–10 weeks post-surgery and later after completion of adjuvant treatment. The aim of the study was to find whether the determination of ctDNA post-surgery and after completion of adjuvant chemotherapy treatment may give information on minimal residual disease, the efficacy of adjuvant treatment, and recurrence in patients with stage III CRC.
On the other hand, ctDNA was detected in 10 out of 66 patients (15%) after finishing treatment with adjuvant chemotherapy. after completion of adjuvant treatment was significantly associated with recurrence free interval (RFI). RFI at 3 years was 30% in patients with detectable ctDNA and 77% in patients with negative ctDNA after chemotherapy (HR 6.8; 95% CI, 11.0–157.0;p< 0.001).
Recently, the same group published a pooled analysis of three cohort studies including 485 stage II–III CRC patients with long term follow-up of 5-years after-surgery ctDNA collection [53]. The authors describe the association of post-surgery ctDNA detection and higher risk of recurrence (38.6% vs. 85.5%;p < 0.001) and poorer OS (64.6% vs. 89.4%;p< 0.001). Furthermore, post-surgery ctDNA status was more accurate in predicting recurrence than individual clinical-pathological risk features such as tumor differentiation, T stage, N stage, lymphovascular invasion, and post-surgery CEA.
All published studies are consistent regarding the clinical impact of ctDNA to detect MRD in stage III CC, not only in identifying patients at high risk of recurrence, but also guiding clinical trials to explore new adjuvant approaches for patients with detectable ctDNA after surgical resection.
Table 1summarizes the main published studies assessing ctDNA prognostic role in localized CC.
| Study | Sample Size | Study Population |
Timepoint of ctDNA Collection |
ctDNA Detection Assay |
Post-op ctDNA Detection Rate |
RFS Post-op ctDNA+ vs. ctDNA− |
|---|---|---|---|---|---|---|
| Tie et al. [43] | 230 | Stage II CC | Weeks 4–10 post-op | Safe-SeqS (1 variant; 15 genes) | 8.7% | 18 (95% CI 7.9–40) p < 0.001 |
| Taieb et al. [33] | 805 | Stage III CC | NA | ddPCR (2 methylated markers) | 13.5% | 1.85 (95% CI 1.31–2.61) p < 0.001 |
| Wang et al. [54] | 58 | Stage I–III CRC | Week 4 post-op | Safe-SeqS (1 variant; 15 genes) | 22.4% | Recurrence-free at 49 months: 33% vs. 100% (non-compared) |
| Reinert et al. [55] | 130 | Stage I–III CRC | Week 4 post-op | Multiplex PCR based NGS assay (SignateraTM) | 10.6% | 7.2 (95% CI 2.7–19.0) p < 0.001 |
| Tie et al. [52] | 96 | Stage III CC (all chemo) | Weeks 4–10 post-op | Safe SeqS (1 variant; 15 genes) | 21% | 3.8 (95% CI 2.4–21.0) p < 0.001 |
| Tie et al. [53] | 485 | Stage II–III CRC and LARC | Weeks 4–10 post-op | Safe-SeqS (1 variant; 15 genes) | 12% | Recurrence-free at 5 years: 38.6% vs. 85.5% p < 0.001 |
| Tarazona et al. [56] | 69 | Stage I–III CC | Weeks 6–8 post-op | ddPCR (2 variants; 29 genes) | 20.3% | 6.96 (95% CI 2.57–18.91) p < 0.001 |
| Scholer et al. [57] | 21 | Stage I–III CRC | Weeks 1–4 post-op | ddPCR (NA) | 28.5% | 37.7 (95% CI 4.2–335.5) p < 0.001 |
CRC: colorectal cancer, CC: colon cancer, NA: non-available, ctDNA: circulating tumor DNA, RFS: recurrence free survival, PCR: polymerase chain reaction, ddPCR: droplet digital PCR, HR: hazard ratio, CI: confidence interval, NGS: next-generation sequencing, Post-op: post-surgery, Pre-op: pre-surgery.
Among patients diagnosed with CRC, 30% have a rectal location [1]. T3/4 or node-positive disease) are usually treated by neoadjuvant CRT or TNT followed by total mesorectal excision (TME) surgery (a mutilating procedure with significant alteration in the quality of life of patients). Moreover, the main site of recurrence in LARC patients is not local but distant metastasis which causes greater morbidity and mortality [58].
Currently, the main prognostic marker in LARC patients is pathological complete response (pCR) as assessed in the rectal surgical specimen after neoadjuvant treatment [59][60].
The need to avoid such mutilating surgery with the associated morbidity has led to an increased interest in the search for prognostic factors to select candidate patients for organ preservation [61]. Currently, watch-and-wait strategies are a possibility in patients with clinical and radiological complete response after TNT. In this setting, the use of ctDNA may potentially help in better identifying patients that are cured after total neoadjuvant therapy and that are potential candidates for a watch-and-wait strategy.
The Australian team led by Tie [62] published a prospective multicenter study that included 159 patients with LARC treated with neoadjuvant CRT followed by TME. ctDNA was detectable in 77%, 8.3%, and 12% of pretreatment, post-CRT, and post-surgery plasma samples, respectively. after CRT or after surgery had a significantly worse RFS irrespective of receiving or not adjuvant CT (HR 6.6; 95% CI 2.6–17;p No association was found between post-CRT ctDNA status and pCR.
In the same year, Khakoo and colleagues [63] investigated the use of ctDNA combined with MRI as an early indicator of response in 47 patients with LARC treated with neoadjuvant CRT followed by TME. Metastatic disease was observed in 70% of patients with positive ctDNA after completing CRT and in 100% of patients with post-surgery positive ctDNA. Furthermore, metastasis-free survival (MFS) was significantly lower in patients with persistent ctDNA
[64] recently published a prospective multicentric study with the aim of analyzing the value of ctDNA in predicting response to neoadjuvant CRT. ctDNA from 104 patients was extracted and analyzed by NGS at four time points: baseline, during neoadjuvant CRT treatment, before surgery, and after surgery. With a median follow-up of 18.8 months, 12.5% of patients developed distant metastases. Moreover, variant allele frequency (VAF) of baseline ctDNA mutations was found to be a significant independent predictor of MFS (HR, 1.27;p< 0.001).
Murahashi et al. [65] studied plasma at baseline and after CRT from 85 patients and found that variations in ctDNA was an independent predictor of complete response to preoperative therapy (p= 0.0276).
Similarly, Pazdirek et al. [66] investigated changes in ctDNA levels of 36 patients with LARC undergoing neoadjuvant CRT and their relationship to treatment response. at baseline was associated with lower DFS and OS at 1.47 and 1.41 years respectively (p= 0.015 andp= 0.010, respectively).
Table 2 summarizes the main published studies assessing ctDNA prognostic role in LARC.
| Study | Sample Size | Study Population |
Timepoint of ctDNA Collection |
ctDNA Detection Assay |
Baseline ctDNA Detection Rate |
Pre-op ctDNA Detection Rate |
Post-op ctDNA Detection Rate |
Main Results for ctDNA+ vs. ctDNA− |
|---|---|---|---|---|---|---|---|---|
| Tie et al. [62] | 159 | LARC | Pre-treatment (CRT), weeks 4–6 post-CRT, and weeks 4–10 post-op | Safe-SeqS (1 variant; 15 genes) | 77% | 8.3% | 12% | Post-CRT RFS: HR 6.6 (95% CI 2.6–17) p < 0.001 Post-op RFS: HR 13 (95% CI 5.5–31) p < 0.001 |
| Khakoo et al. [63] | 47 | Localized rectal cancer | Pre-treatment (CRT), mid-CRT, post-CRT, and weeks 4–12 post-op | ddPCR (up to 3 variants; 6 genes) | 74% | 21% | 13% | Post-CRT MFS: 7.1 (95% CI 2.4–21.5) p < 0.001 Post-op DFS: 39.9 (95% CI 4.0–399.5) p = 0.002 |
| Zhou et al. [64] | 104 | LARC | Pre-treatment (CRT), 1 week from the start of treatment, post-CRT, and 4 weeks post-op | HiSeq 3000 Sequencing System (IlluminaTM). Panel of 1021 genes |
75% | 10.5% | 6.7% | Post-CRT MFS:19.82 (95% CI 2.029–193.7) p < 0.001 Post-op MFS: 25.30 (95% CI, 1.475–434) p < 0.001 |
| Murahashi et al. [65] | 85 | LARC | Pre-treatment (CRT), post-CRT, and 12 weeks post-op | Oncomine CRC (14 genes) | 57.6% | 22.3% | NA | Post-op RFS: 17.1 (95% CI, 1.0–282) p < 0.001 |
| Pazdirek et al. [66] | 36 | LARC | Pre-treatment (CRT), 1 week from the start of treatment | Denaturing capillary electrophoresis (DCE) and High sensitivity Beaming assay | 21.2% | NA | NA | Prior CRT: reduction DFS by 1.47 years (p = 0.015) and OS by 1.41 years (p = 0.010) |
| Vidal et al. [67] | 62 | LARC | Pre-treatment (TNT) and post-CRT (48 h pre-op) | LUNAR-1 | 83% | 15% | NA | Post-CRT RFS: HR 4.029 (95% CI, 1.004–16.16) p = 0.033 Post-CRT OS: HR 23(95% CI, 2.4–212) p < 0.0001 |
LARC: locally advanced rectal cancer, CRC: colorectal cancer, CRT: chemo-radiotherapy, TNT: total neoadjuvant treatment, MFS: metastasis free-survival, ctDNA: circulating tumor DNA, RFS: recurrence free survival, ddPCR: droplet digital PCR, HR: hazard ratio, CI: confidence interval, Post-op: post-surgery, Pre-op: pre-surgery.
following TNT and before surgery in patients that are likely to recur at distant sites. Plasma samples were collected at baseline and after TNT within 48 h before surgery (pre-surgery). Patients with pre-surgery positive ctDNA had an increased risk of recurrence compared to patients with negative ctDNA In line with previous works in LARC, no correlation was found between pre-surgery ctDNA
Globally, in LARC there are three timepoints where ctDNA has been mostly interrogated: baseline, after neoadjuvant treatment, and post-surgery. The prognostic role of post-surgery ctDNA is consistent with previous studies in CC and confirms the value of ctDNA to detect MRD. Finally, pre-surgery ctDNA analysis after neoadjuvant treatment (either CRT or TNT) detects minimal metastatic disease (MMD), systemic recurrence and death. Prospective clinical trials are needed to validate these finding by assessing systemic treatment intensification in pre-surgery ctDNA positive patients.