The MYB binding protein 1A (MYBBP1A, also known as p160) acts as a co-repressor of multiple transcription factors involved in many physiological processes. Therefore, MYBBP1A acts as a tumor suppressor in multiple aspects related to cell physiology, most of them very relevant for tumorigenesis. We explored the different roles of MYBBP1A in different aspects of cancer, such as mitosis, cellular senescence, epigenetic regulation, cell cycle, metabolism plasticity and stemness. We especially reviewed the relationships between MYBBP1A, the inhibitory role it plays by binding and inactivating c-MYB and its regulation of PGC-1α, leading to an increase in the stemness and the tumor stem cell population. In addition, MYBBP1A causes the activation of PGC-1α directly and indirectly through c-MYB, inducing the metabolic change from glycolysis to oxidative phosphorylation (OXPHOS). Therefore, the combination of these two effects caused by the decreased expression of MYBBP1A provides a selective advantage to tumor cells. Interestingly, this only occurs in cells lacking pVHL. Finally, the loss of MYBBP1A occurs in 8%–9% of renal tumors. tumors, and this subpopulation could be studied as a possible target of therapies using inhibitors of mitochondrial respiration.
1. Introduction
MYBBP1A is a ubiquitous 160 kDa protein, first identified for its binding and repression of the proto-oncogene c-MYB. Since then, it has been found that MYBBP1A acts as a co-repressor of multiple transcription factors involved in many important physiological processes. In all these processes, MYBBP1A acts as a tumor suppressor, regulating the evolution and malignity of cells. Furthermore, the loss of MYBBP1A occurs in 8%–9% of renal tumors and may occur in a percentage of other tumors, according to its role as a potent tumor suppressor. However, to date, there has been no systematic review of the roles of MYBBP1A, and how it may control these processes. And more importantly, there is no thorough review of how the loss of MYBBP1A contributes to the development and evolution of tumors. It also will be interesting to explore whether this subpopulation could be studied as a possible target of new therapies.
2. Tumor Suppressor Roles of MYBBP1A
2.1. MYB Binding Protein 1A
The MYB binding protein 1A (MYBBP1A, also known as p160) is a 160 kDa protein that is expressed in all tissues. It is located predominantly in the nucleolus, although its presence in the nucleoplasm has also been observed
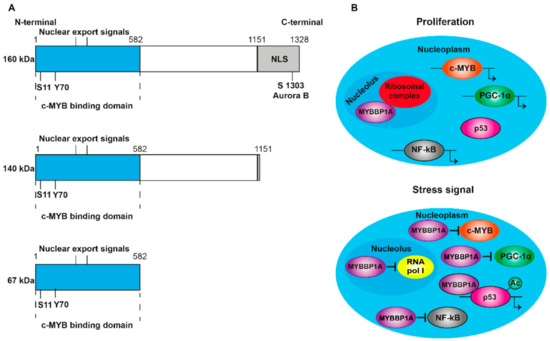
[1]. It contains nuclear and nucleolar localization sequences (NLS) at the C-terminal end (A) and is anchored to the nucleolus through its interaction with RNA. MYBBP1A translocates from the nucleolus to the nucleoplasm when there is a decrease in the RNA content caused by stress signals, such as the absence of glucose or the inhibition of ribosome synthesis
[2][3]. In addition, ribosomal stress induces the processing of MYBBP1A in its shortened forms (140 and 67 kDa) (A), which are found in the nucleoplasm following the loss of the nucleolar localization signal. Although p160 is expressed in al tissues, the 67 kDa protein is only expressed in a few subcultivated cell lines. MYBBP1A is a highly evolutionarily conserved protein, presenting some homology with the Pol5p protein of Saccharomyces cerevisiae, and is involved in the production of ribosomal RNA (rRNA)
[4]. Initially, it was identified as a protein that was bound to the protein encoded by the proto-oncogene c-MYB
[1], and, subsequently, its binding to numerous transcription factors has been discovered. For this reason, it is thought that the function of MYBBP1A is the regulation of the activity of several transcription factors, acting as a regulator of fundamental biological processes such as cell division, proliferation and apoptosis.
Figure 1. Structure and translocation model of MYBBP1A. (A) Scheme of the domains and phosphorylation sites of MYBBP1A and their shortened forms, produced by proteolytic cleavage. NLS: nuclear and nucleolar localization sequence. S1303 Aurora B indicates that MYBBP1A is phosphorylated by Aurora B kinase at serine residue 1303. ARNpolI is RNA polymerase I. (B) Model, proposed by Hochstatter et al., suggests that MYBBP1A is located in the nucleolus of the proliferative cells attached to the ribosomal complex. However, in cells subjected to some stress signals, such as glucose deprivation, MYBBP1A translocates mainly to the nucleoplasm, where it binds to different transcription factors to interrupt cell cycle progression, proliferation and energy production.
Mori et al.
[5] deleted the
mybbp1a gene in mice and obtained
mybbp1a +/- heterozygous healthy and fertile mice but failed to generate any
mybbp1a -/- mice. After analyzing the embryos in depth at different points of development (days 6.5, 9.5 and 11.5), they found no homozygous
mybbp1a -/- embryos. They also isolated blastocysts from heterozygous mice, kept them in culture for several days and genotyped them without detecting any
mybbp1a -/- blastocysts. Therefore, although the molecular mechanism involved is unclear,
mybbp1a is essential in early embryonic development.
2.2. Regulation of MYBBP1A
MYBBP1A is a ubiquitination target of the von Hippel–Lindau tumor suppressor gene (VHL)
[6], which is a component of the E3 ubiquitin ligase complex. The loss of function mutations in this gene is associated with von Hippel–Lindau disease, a hereditary cancer syndrome characterized by a high risk of developing highly vascularized tumors in the eyes, brain and spine, as well as benign or malignant tumors in the kidney, pancreas and adrenal medulla
[7]. The biallelic inactivation of VHL is also frequent in renal carcinomas and sporadic hemangioblastomas
[7][8]. It has been found that the VHL protein (pVHL) binds directly to MYBBP1A, causing its degradation in an iron- and proteasome-dependent manner
[6]. The region of MYBBP1A, located between amino acids 632 and 694, is necessary for this degradation to occur. This region contains a sequence similar to the one surrounding the hydroxylated proline in the HIF1α protein that is required for its degradation, which also occurs via pVHL. It has also been found that the mutation in the proline residue 693 of MYBBP1A (Pro693), when changed to alanine, blocks the degradation of MYBBP1A by pVHL. Moreover, as in HIF1α degradation, MYBBP1A degradation by pVHL also requires iron, as iron chelation with desferrioxamine (DFO) abolishes it. Based on these findings, it has been proposed that the Pro693 residue of MYBBP1A is hydroxylated in the presence of iron and oxygen, leading to its ubiquitination by pVHL and its subsequent degradation via the proteasome. Despite the similarities between MYBBP1A and HIF1α degradation by pVHL in terms of amino acid sequence and iron requirement, how hypoxia affects MYBBP1A’s stability at protein and mRNA levels is still unknown.
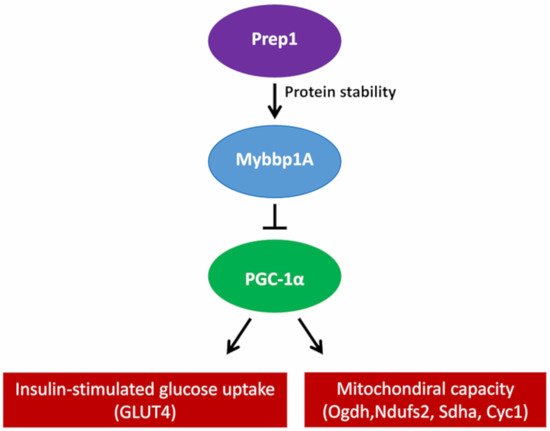
However, pVHL is not the only protein involved in the regulation of MYBBP1A. PREP1 controls the half-life of MYBBP1A at the protein stability level, since the interaction between PREP1 and MYBBP1A prevents its degradation via the proteasome. The muscles of hypomorphic mice for Prep1 show reduced levels of Mybbp1A, but an increase in the peroxisome proliferator-activated receptor γ coactivator (Pgc-1α), a well-known regulator of mitochondrial metabolism and biogenesis. Pgc-1α upregulation occurs at both the mRNA and protein levels, accompanied by an increase in the expression of the Glut4 transporter. Conversely, this effect is revoked by the direct delivery of Mybbp1A cDNA in the muscle of Prep1-deficient mice, decreasing Pgc-1α and Glut4 expression
[9]. Furthermore, the elimination of Prep1 specifically in the muscles of mice causes an increase in the expression of the genes of the respiratory chain and Pgc-1α, together with a significant reduction in the levels of Mybbp1a ()
[10].
Figure 2. Proposed mechanism of how Prep1 regulates Mybbp1a stability, modulating Pgc-1α, mitochondrial genes and Glut 4 expression in mouse skeletal muscle.
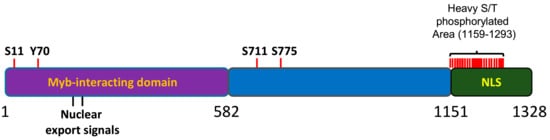
MYBBP1A has been found to be posttranslationally modified; however, the biological relevance of these modifications has not yet been clarified. Importantly, the MYBBP1A protein is heavily phosphorylated in cells
[11][12][13][14]. Most of the sites for phosphorylation (18 out of 21) are mapped to the C terminal region of the protein, within a 200 aa portion that has been found to be relevant for the nucleolar localization of the protein
[15] (). Accordingly, MYBBP1A was found to be an important component of the phosphoproteome of the mitotic spindle; however, the Aurora B kinase is the only proven kinase acting on the protein in vitro
[16]. MYBBP1A can be phosphorylated by different kinases, but scarce data have been shown for many of them.
Figure 3. Schematic representation of MYBBP1A protein. 1-582aa domain, reported to interact with the Myb protein. NLS: nuclear and nucleolar localization signal. S, T or Y are the phophorylation residues reported in several phosphoproteomic assays. Adapted from the atlas of genetics and cytogenetics in oncology and haematology.
2.3. MYBBP1A as a Regulator of Transcription Factors
MYBBP1A was first identified because of its binding to the proto-oncogene c-MYB
[1]. The proto-oncogene c-MYB codes for the transcription factor c-MYB, which is an essential regulator of stem cells and progenitors of the bone marrow, colon crypts and neurogenic regions of the adult brain
[17]. Its main function is to maintain the proliferative state and the immature characteristics of these cells. The oncogenic forms of this protein are generated by loss of the N-terminus, C-terminus or both, maintaining the DNA binding domain and the region necessary for the transactivation of transcription. At the C-terminal end is the negative regulatory domain (NRD), which negatively regulates transactivation and the binding of the protein to DNA
[18][19]. Both MYBBP1A and its shortened 67 kDa form, which are generated by proteolytic cleavage in some cell types, bind to the NRD of c-MYB. This binding occurs through the leucine zipper motifs present in the N-terminal fragment of MYBBP1A and in the NRD of c-MYB
[1].
It has also been found that MYBBP1A acts as a repressor of PGC-1α. Both the N-terminal and the C-terminal domain of MYBBP1A bind to the negative regulatory domain of PGC-1α and reduce its ability to stimulate mitochondrial respiration and the expression of electron transport chain genes. This negative regulation is inhibited when PGC-1α is phosphorylated by the p38 MAP kinase
[20]. Another transcription factor regulated by MYBBP1A is PREP1
[21]. Both the 160 kDa and the 67 kDa version of MYBBP1A bind to the first homology domain (HR1) of PREP1, inhibiting its transcriptional activity and the expression of the
HoxB2 gene in NT2-D1 cells treated with retinoic acid. MYBBP1A competes with PBX1 to bind to PREP1
[21]. MYBBP1A also competes with p300 for binding to the RelA/p65 subunit of NF-κB, acting as a repressor of its activity when the binding occurs
[22]. In addition, MYBBP1A acts as a corepressor of CRY1 in the period2 promoter, inhibiting the expression of this gene, which is involved in the regulation of the circadian clock
[23]. Despite its general role as a negative regulator of transcription factors, MYBBP1A also acts as an activator of some transcription factors, such as the aromatic hydrocarbon receptor (AhR)
[24].
2.4. MYBBP1A and Epigenetic Regulation
MYBBP1A also binds to factors involved in epigenetic regulation. It has been found that MYBBP1A is a component of the ATP-dependent chromatin remodeling complex (esBAF), which is associated with differentiation processes and stem cell phenotypes. Although the role of MYBBP1A in this complex is not clear, it could be related to epigenetic regulatory functions
[25][26]. Likewise, it has been shown that MYBBP1A acts as a regulator of muscle-specific gene expression and the differentiation of myoblasts by binding to MyoD. The binding of MYBBP1A and MyoD in certain regions of genes that are not expressed in muscle tissue decreases the transcriptional activity of MyoD. It has been proposed that MYBBP1A carries out this repressive function by recruiting negative epigenetic modifiers, such as Histone Deacetylase 1 and 2, HDAC1/2 and Suv39h1, inducing a less accessible structure of chromatin
[27].
It has also been found that MYBBP1A plays an important role in the acetylation and activation of p53
[2][28][29][30][31][32]. Under stress conditions in the nucleolus, which are produced by the inhibition of the ribosome synthesis or by the absence of glucose, rRNA transcription is blocked, and the RNA content in the nucleolus is reduced. This decrease causes the translocation of MYBBP1A to the nucleoplasm, where it facilitates the interaction between p53 and p300, increasing the acetylation of p53. Specifically, the proposed model states that stress in the nucleolus causes the inactivation of Human Double Minute 2, HDM2, which leads to the stabilization of p53 and the formation of non-acetylated p53 dimers. Once translocated to the nucleoplasm, MYBBP1A binds to these p53 dimers through its central region (aa 643–1150) and C-terminal end (aa 1151–1328). After this interaction occurs, dimers are formed between units of MYBBP1A bound to p53 dimers, resulting in the formation of p53 tetramers. The transcription coactivator p300 binds to these structures, acetylates the p53 units, and then MYBBP1A dissociates from the complex. Both MYBBP1A-p53 binding sites (aa 643–1150 and aa 1151–1328) are required to facilitate p53 tetramerization and acetylation, so that only full-length MYBBP1A is able to promote p53 activation. Finally, the tetramer of acetylated p53, still bound to p300, binds to the promoters of its target genes and induces the expression of genes related to apoptosis and/or cell cycle arrest
[31].
2.5. Role of MYBBP1A in Cell Cycle and Mitosis
MYBBP1A has been linked to cell cycle control, especially the G2/M phase, and mitosis. On the one hand, MYBBP1A silencing in HeLa cells induces the up-regulation of genes that inhibit growth, such as BRCA1, TOP2A and WEE1, and the down-regulation of genes involved in DNA repair, such as CDKN1a and GADD45b, ultimately provoking a cell cycle arrest at G2/M
[5]. On the other hand, there are several findings suggesting that MYBBP1A participates in the regulation of mitosis. It has been found that MYBBP1A translocates from the nucleolus to the nucleoplasm during mitosis, situating itself in parachromosomal regions in metaphase–anaphase, where it colocalizes with the chromatin marker phosphohistone H3
[5]. In addition, MYBBP1A is phosphorylated by Aurora B kinase at serine residue 1303. The peak of this phosphorylation occurs at the G2/M transition and is completely inhibited when cells are treated with danusertib, an inhibitor of Aurora kinases. This phosphorylation of MYBBP1A in G2/M is performed exclusively by the Aurora B kinase, since only the silencing of Aurora B (and not A or C) suppresses this phosphorylation. It has also been proposed that MYBBP1A is part of a set of proteins involved in the progression of mitosis, since the reduction in MYBBP1A expression caused by RNA interference (RNAi) causes both a delay in the progression of mitosis and failures in the assembly and stability of the mitotic spindle
[16].
During mitosis, the nucleus breaks down, and the nucleolar proteins are translocated to the cytoplasm. Among these proteins is MYBBP1A, which, as mentioned, induces the acetylation of p53, and this interaction is essential at the G1 postmitotic control point. In this context, MYBBP1A participates in the activation of the control point machinery when mitosis is abnormally prolonged
[33].