Flowering is one of the most critical developmental transitions in plants’ life. The irreversible change from the vegetative to the reproductive stage is strictly controlled to ensure the progeny’s success. In Arabidopsis thaliana, seven flowering genetic pathways have been described under specific growth conditions. However, the evidence suggests that these pathways are tightly interconnected in a complex multilevel regulatory network. Here we summarized the information of our recent publication.
- flowering transition
- genetic regulatory network
- multilevel regulation
1. Introduction
2. Regulation of Flowering Repressors
3. Endogenous Signals in Flowering Transition
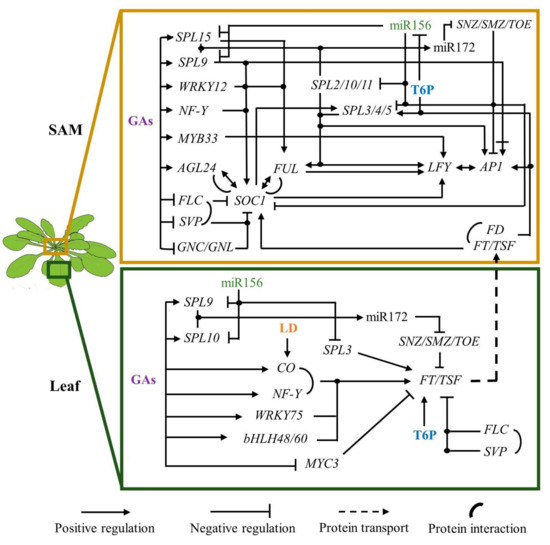
4. Long-Day Photoperiod and High-Temperature Function as Inductive Signals
Day length and high permissible temperatures are important signals to induce flowering in A. thaliana. Detection of photoperiod relies primarily on CO protein, a B-box-type zinc finger TF with a CCT domain, that accumulates during the day in the vascular tissue [4]. CONSTITUTTIVE PHOTOMORFOGENIC 1 (COP1) and SUPPRESSOR OF PHYA-105S 1 (COP1/SPA1) complex ubiquitinates CO protein to be degraded by the proteasome at night. During the diurnal phase of long-day (LD)-photoperiod, cryptochromes 1 and 2, and phytochrome A are activated by blue and far-red light, respectively, inhibiting COP1/SPA1 activity. This action allows CO accumulation in the afternoon [65][66].
CO induces FT expression [18] which is translated and systemically travels from rosette leaves to the SAM to induce flowering [67][68]. Once they reach the SAM, FT interacts with the bZIP transcription factor FD via the 14-3-3 growth response factors [69]. Moreover, the FT-FD complex regulates the expression of SPL3/4/5 in the IM, and they directly or in association with FD upregulate FUL, LFY, and AP1 [70][71].
PHYTOCHROME INTERACTING FACTOR 4 (PIF 4) and its orthologs PIF5 and PIF7 are responsible for inducing FT and TSF expression in response to high temperature under short-day conditions [72][73]. Interestingly, at 27 °C, the H2A.Z-nucleosomes levels decreased at the FT locus, relaxing the chromatin and favoring the union of PIF4 to FT DNA [72]. It seems, therefore, that LD-photoperiod and high temperature are signals that can be separated, however, more research is required to establish their possible synergism.
The actual hierarchical flowering model proposed that different inputs converge into the integrators that transduce these signals to the FM identity genes [1]. Alternatively, we propose that endogenous signals can transversally dictate whether the plants remain in the vegetative phase or initiate the reproductive state. In this view, miR156, the DELLA proteins, and possibly low concentration of certain carbohydrates maintain the vegetative state, while miR172, GA, and T6P allow the reproductive phase change. Inductive signals like LD-photoperiod and temperature accelerate the flowering transition process in plants such as Arabidopsis, in part, by upregulating FT in the leaves and a group of the SPLs and MADS-box genes in the apical meristem. This ensures that flowering happens when the external conditions are optimal for those species.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22115716
References
- Miguel A. Blázquez; Cristina Ferrándiz; Francisco Madueño; François Parcy; How Floral Meristems are Built. Plant Molecular Biology 2006, 60, 855-870, 10.1007/s11103-006-0013-z.
- Enrico S. Coen; Elliot M. Meyerowitz; The war of the whorls: genetic interactions controlling flower development. Nature 1991, 353, 31-37, 10.1038/353031a0.
- Israel Ausin; Carlos Alonso-Blanco; Jose-Miguel Martinez-Zapater; Environmental regulation of flowering. The International Journal of Developmental Biology 2004, 49, 689-705, 10.1387/ijdb.052022ia.
- Joanna Putterill; Frances Robson; Karen Lee; Rüdiger Simon; George Coupland; The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847-857, 10.1016/0092-8674(95)90288-0.
- Paula Suárez-López; Kay Wheatley; Frances Robson; Hitoshi Onouchi; Federico Valverde; George Coupland; CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116-1120, 10.1038/35074138.
- Takato Imaizumi; Steve A. Kay; Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science 2006, 11, 550-558, 10.1016/j.tplants.2006.09.004.
- Miguel A. Blazquez; Roland Green; Ove Nilsson; Michael R. Sussman; Detlef Weigel; Gibberellins Promote Flowering of Arabidopsis by Activating the LEAFY Promoter. The Plant Cell 1998, 10, 791, 10.2307/3870665.
- Aimone Porri; Stefano Torti; Maida Romera-Branchat; George Coupland; Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198-2209, 10.1242/dev.077164.
- Candice C. Sheldon; Dean T. Rouse; E. Jean Finnegan; W. James Peacock; Elizabeth S. Dennis; The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences 2000, 97, 3753-3758, 10.1073/pnas.060023597.
- Gordon G Simpson; The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology 2004, 7, 570-574, 10.1016/j.pbi.2004.07.002.
- M Koornneef; C Alonso-Blanco; H Blankestijn-De Vries; C J Hanhart; A J Peeters; Genetic interactions among late-flowering mutants of Arabidopsis.. Genetics 1998, 148, 885-892.
- Miguel Blazquez; Ji Hoon Ahn; Detlef Weigel; A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics 2003, 33, 168-171, 10.1038/ng1085.
- Jeong Hwan Lee; Seong Jeon Yoo; Soo Hyun Park; Ildoo Hwang; Ji Hoon Ahn; Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes & Development 2007, 21, 397-402, 10.1101/gad.1518407.
- Gang Wu; R. Scott Poethig; Temporal regulation of shoot development in Arabidopsis thalianaby miR156 and its target SPL3. Development 2006, 133, 3539-3547, 10.1242/dev.02521.
- Youbong Hyun; René Richter; George Coupland; Competence to Flower: Age-Controlled Sensitivity to Environmental Cues. Plant Physiology 2017, 173, 36-46, 10.1104/pp.16.01523.
- Vanessa Wahl; Jathish Ponnu; Armin Schlereth; Stéphanie Arrivault; Tobias Langenecker; Annika Franke; Regina Feil; John E. Lunn; Mark Stitt; Markus Schmid; et al. Regulation of Flowering by Trehalose-6-Phosphate Signaling in Arabidopsis thaliana. Science 2013, 339, 704-707, 10.1126/science.1230406.
- Horim Lee; Sung-Suk Suh; Eunsook Park; Euna Cho; Ji Hoon Ahn; Sang-Gu Kim; Jong Seob Lee; Young Myung Kwon; Ilha Lee; The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes & Development 2000, 14, 2366-2376, 10.1101/gad.813600.
- Alon Samach; Hitoshi Onouchi; Scott E. Gold; Gary S. Ditta; Zsuzsanna Schwarz-Sommer; Martin F. Yanofsky; George Coupland; Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science 2000, 288, 1613-1616, 10.1126/science.288.5471.1613.
- Jihyun Moon; Horim Lee; Minsoo Kim; Ilha Lee; Analysis of Flowering Pathway Integrators in Arabidopsis. Plant and Cell Physiology 2005, 46, 292-299, 10.1093/pcp/pci024.
- Scott D. Michaels; Edward Himelblau; Sang Yeol Kim; Fritz M. Schomburg; Richard M. Amasino; Integration of Flowering Signals in Winter-Annual Arabidopsis. Plant Physiology 2005, 137, 149-156, 10.1104/pp.104.052811.
- F.D. Hempel; D. Weigel; M.A. Mandel; G. Ditta; P.C. Zambryski; L.J. Feldman; M.F. Yanofsky; Floral determination and expression of floral regulatory genes in Arabidopsis. Development 1997, 124, 3845-3853, 10.1242/dev.124.19.3845.
- Frédéric Bouché; Guillaume Lobet; Pierre Tocquin; Claire Périlleux; FLOR-ID: an interactive database of flowering-time gene networks inArabidopsis thaliana. Nucleic Acids Research 2016, 44, D1167-D1171, 10.1093/nar/gkv1054.
- Giorgio Perrella; Elisa Vellutini; Anna Zioutopoulou; Eirini Patitaki; Lauren R. Headland; Eirini Kaiserli; Let it bloom: cross‐talk between light and flowering signaling in Arabidopsis. Physiologia Plantarum 2020, 169, 301-311, 10.1111/ppl.13073.
- Candice C. Sheldon; Joanne E. Burn; Pascual P. Perez; Jim Metzger; Jennifer A. Edwards; W. James Peacock; Elizabeth S. Dennis; The FLF MADS Box Gene: A Repressor of Flowering in Arabidopsis Regulated by Vernalization and Methylation. The Plant Cell 1999, 11, 445, 10.2307/3870872.
- Ulrike Hartmann; Susanne Hohmann; Klaus Nettesheim; Ellen Wisman; Heinz Saedler; Peter Huijser; Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. The Plant Journal 2000, 21, 351-360, 10.1046/j.1365-313x.2000.00682.x.
- Katia C. Scortecci; Scott D. Michaels; Richard M. Amasino; Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. The Plant Journal 2001, 26, 229-236, 10.1046/j.1365-313x.2001.01024.x.
- Weiwei Deng; Hua Ying; Chris A. Helliwell; Jen Taylor; W. James Peacock; Elizabeth S. Dennis; FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences 2011, 108, 6680-6685, 10.1073/pnas.1103175108.
- Chris A. Helliwell; Craig C. Wood; Masumi Robertson; W. James Peacock; Elizabeth S. Dennis; The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 2006, 46, 183-192, 10.1111/j.1365-313x.2006.02686.x.
- Csaba Hornyik; Céline Duc; Katarzyna Rataj; Lionel C. Terzi; Gordon G. Simpson; Alternative polyadenylation of antisense RNAs and flowering time control. Biochemical Society Transactions 2010, 38, 1077-1081, 10.1042/bst0381077.
- Sebastian Marquardt; Oleg Raitskin; Zhe Wu; Fuquan Liu; Qianwen Sun; Caroline Dean; Functional Consequences of Splicing of the Antisense Transcript COOLAIR on FLC Transcription. Molecular Cell 2014, 54, 156-165, 10.1016/j.molcel.2014.03.026.
- Zhe Wu; Xiaofeng Fang; Danling Zhu; Caroline Dean; Autonomous Pathway: FLOWERING LOCUS C Repression through an Antisense-Mediated Chromatin-Silencing Mechanism. Plant Physiology 2020, 182, 27-37, 10.1104/pp.19.01009.
- Yongke Tian; Han Zheng; Fei Zhang; Shiliang Wang; Xiaoru Ji; Chao Xu; Yuehui He; Yong Ding; PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Science Advances 2019, 5, eaau7246, 10.1126/sciadv.aau7246.
- Scott Berry; Caroline Dean; Environmental perception and epigenetic memory: mechanistic insight through FLC. The Plant Journal 2015, 83, 133-148, 10.1111/tpj.12869.
- Ulrich Lutz; Thomas Nussbaumer; Manuel Spannagl; Julia Diener; Klaus Fx Mayer; Claus Schwechheimer; Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis. eLife 2017, 6, e22114, 10.7554/elife.22114.
- David Posé; Leonie Verhage; Felix Ott; Levi Yant; Johannes Mathieu; Gerco C. Angenent; Richard Immink; Markus Schmid; Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414-417, 10.1038/nature12633.
- Keh Chien Lee; Kyung Sook Chung; Hee Tae Lee; Jae-Hyeok Park; Jeong Hwan Lee; Jeong-Kook Kim; Role of Arabidopsis Splicing factor SF1 in Temperature-Responsive Alternative Splicing of FLM pre-mRNA. Frontiers in Plant Science 2020, 11, 1-14, 10.3389/fpls.2020.596354.
- Giovanna Capovilla; Efthymia Symeonidi; Rui Wu; Markus Schmid; Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. Journal of Experimental Botany 2017, 68, 5117-5127, 10.1093/jxb/erx328.
- Gang Wu; Mee Yeon Park; Susan R. Conway; Jia-Wei Wang; Detlef Weigel; R. Scott Poethig; The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell 2009, 138, 750-759, 10.1016/j.cell.2009.06.031.
- Youbong Hyun; René Richter; Coral Vincent; Rafael Martinez-Gallegos; Aimone Porri; George Coupland; Multi-layered Regulation of SPL15 and Cooperation with SOC1 Integrate Endogenous Flowering Pathways at the Arabidopsis Shoot Meristem. Developmental Cell 2016, 37, 254-266, 10.1016/j.devcel.2016.04.001.
- Jae-Hoon Jung; Pil Joon Seo; Seok Ki Kang; Chung-Mo Park; miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Molecular Biology 2011, 76, 35-45, 10.1007/s11103-011-9759-z.
- A. Lang; THE EFFECT OF GIBBERELLIN UPON FLOWER FORMATION. Proceedings of the National Academy of Sciences 1957, 43, 709-717, 10.1073/pnas.43.8.709.
- Sven Eriksson; Henrik Böhlenius; Thomas Moritz; Ove Nilsson; GA4 Is the Active Gibberellin in the Regulation of LEAFY Transcription and Arabidopsis Floral Initiation. The Plant Cell 2006, 18, 2172-2181, 10.1105/tpc.106.042317.
- Jae-Hoon Jung; Yun Ju; Pil Joon Seo; Jae-Hyung Lee; Chung-Mo Park; The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. The Plant Journal 2012, 69, 577-588, 10.1111/j.1365-313x.2011.04813.x.
- Johannes Mathieu; Levi Yant; Felix Mürdter; Frank Küttner; Markus Schmid; Repression of Flowering by the miR172 Target SMZ. PLOS Biology 2009, 7, e1000148, 10.1371/journal.pbio.1000148.
- Nobutoshi Yamaguchi; Cara M. Winter; Miin-Feng Wu; Yuri Kanno; Ayako Yamaguchi; Mitsunori Seo; Doris Wagner; Gibberellin Acts Positively Then Negatively to Control Onset of Flower Formation in Arabidopsis. Science 2014, 344, 638-641, 10.1126/science.1250498.
- Mingzhe Li; Fengying An; Wenyang Li; Mengdi Ma; Ying Feng; Xing Zhang; Hongwei Guo; DELLA proteins interact with FLC to repress flowering transition. Journal of Integrative Plant Biology 2016, 58, 642-655, 10.1111/jipb.12451.
- Xingliang Hou; Jiannan Zhou; Chang Liu; Lu Liu; Lisha Shen; Hao Yu; Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nature Communications 2014, 5, 4601, 10.1038/ncomms5601.
- René Richter; Emmanouil Bastakis; Claus Schwechheimer; Cross-Repressive Interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the Control of Greening, Cold Tolerance, and Flowering Time in Arabidopsis. Plant Physiology 2013, 162, 1992-2004, 10.1104/pp.113.219238.
- Yanchong Yu; Zhenhua Liu; Long Wang; Sang-Gyu Kim; Pil J. Seo; Meng Qiao; Nan Wang; Shuo Li; Xiaofeng Cao; Chung-Mo Park; et al.FengNing Xiang WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. The Plant Journal 2016, 85, 96-106, 10.1111/tpj.13092.
- Liping Zhang; Ligang Chen; Diqiu Yu; Transcription Factor WRKY75 Interacts with DELLA Proteins to Affect Flowering. Plant Physiology 2018, 176, 790-803, 10.1104/pp.17.00657.
- Zhenbing Ma; Wei Li; Houping Wang; Diqiu Yu; WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age‐mediated flowering. Journal of Integrative Plant Biology 2020, 62, 1659-1673, 10.1111/jipb.12946.
- Wei Li; Houping Wang; Diqiu Yu; Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Molecular Plant 2016, 9, 1492-1503, 10.1016/j.molp.2016.08.003.
- Vicente Balanzà; Irene Martínez-Fernández; Cristina Ferrándiz; Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. Journal of Experimental Botany 2014, 65, 1193-1203, 10.1093/jxb/ert482.
- Fernando Andrés; Aimone Porri; Stefano Torti; Julieta Mateos; Maida Romera-Branchat; José Luis García-Martínez; Fabio Fornara; Veronica Gregis; Martin M. Kater; George Coupland; et al. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proceedings of the National Academy of Sciences 2014, 111, E2760-E2769, 10.1073/pnas.1409567111.
- Chang Liu; Hongyan Chen; Hong Ling Er; Hui Meng Soo; Prakash P. Kumar; Jin-Hua Han; Yih Cherng Liou; Hao Yu; Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481-1491, 10.1242/dev.020255.
- Rigoberto V. Pérez-Ruiz; Berenice García-Ponce; Nayelli Marsch Martinez; Yamel Ugartechea-Chirino; Mitzi Villajuana-Bonequi; Stefan de Folter; Eugenio Azpeitia; José Dávila-Velderrain; David Cruz-Sánchez; Adriana Garay; et al.María De La Paz SanchezJuan M. Estévez-PalmasElena R. Álvarez-Buylla XAANTAL2 (AGL14) Is an Important Component of the Complex Gene Regulatory Network that Underlies Arabidopsis Shoot Apical Meristem Transitions. Molecular Plant 2015, 8, 796-813, 10.1016/j.molp.2015.01.017.
- Jungeun Lee; Mijin Oh; Hanna Park; Ilha Lee; SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. The Plant Journal 2008, 55, 832-843, 10.1111/j.1365-313x.2008.03552.x.
- Gregory F.W. Gocal; Candice C. Sheldon; Frank Gubler; Thomas Moritz; David J. Bagnall; Colleen P. Macmillan; Song F. Li; Roger W. Parish; Elizabeth S. Dennis; Detlef Weigel; et al.Rod W. King GAMYB-like Genes, Flowering, and Gibberellin Signaling in Arabidopsis. Plant Physiology 2001, 127, 1682-1693, 10.1104/pp.127.4.1682.
- Miguel Blazquez; Detlef Weigel; Integration of floral inductive signals in Arabidopsis. Nature 2000, 404, 889-892, 10.1038/35009125.
- Laurent Corbesier; Pierre Lejeune; Georges Bernier; The role of carbohydrates in the induction of flowering in Arabidopsis thaliana : comparison between the wild type and a starchless mutant. Planta 1998, 206, 131-137, 10.1007/s004250050383.
- Matthew J. Paul; Lucia F. Primavesi; Deveraj Jhurreea; Yuhua Zhang; Trehalose Metabolism and Signaling. Annual Review of Plant Biology 2008, 59, 417-441, 10.1146/annurev.arplant.59.032607.092945.
- Manuel Buendía-Monreal; C. Stewart Gillmor; Convergent repression of miR156 by sugar and the CDK8 module of Arabidopsis Mediator. Developmental Biology 2017, 423, 19-23, 10.1016/j.ydbio.2017.01.007.
- Hang Zhao; Ke Lin; Lin Ma; Qingshuai Chen; Shuo Gan; Gang Li; Arabidopsis NUCLEAR FACTOR Y A8 inhibits the juvenile-to-adult transition by activating transcription of MIR156s. Journal of Experimental Botany 2020, 71, 4890-4902, 10.1093/jxb/eraa197.
- René Richter; Atsuko Kinoshita; Coral Vincent; Rafael Martinez-Gallegos; He Gao; Annabel D. Van Driel; Youbong Hyun; Julieta L. Mateos; George Coupland; Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLOS Genetics 2019, 15, e1008065, 10.1371/journal.pgen.1008065.
- Zecheng Zuo; Hongtao Liu; Bin Liu; Xuanming Liu; Chentao Lin; Blue Light-Dependent Interaction of CRY2 with SPA1 Regulates COP1 activity and Floral Initiation in Arabidopsis. Current Biology 2011, 21, 841-847, 10.1016/j.cub.2011.03.048.
- Li-Jun Liu; Yan-Chun Zhang; Qing-Hua Li; Yi Sang; Jian Mao; Hong-Li Lian; Long Wang; Hong-Quan Yang; COP1-Mediated Ubiquitination of CONSTANS Is Implicated in Cryptochrome Regulation of Flowering in Arabidopsis. The Plant Cell 2008, 20, 292-306, 10.1105/tpc.107.057281.
- Laurent Corbesier; Coral Vincent; Seonghoe Jang; Fabio Fornara; Qingzhi Fan; Iain Searle; Antonis Giakountis; Sara Farrona; Lionel Gissot; Colin Turnbull; et al.George Coupland FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030-1033, 10.1126/science.1141752.
- Katja E. Jaeger; Philip A. Wigge; FT Protein Acts as a Long-Range Signal in Arabidopsis. Current Biology 2007, 17, 1050-1054, 10.1016/j.cub.2007.05.008.
- Silvio Collani; Manuela Neumann; Levi Yant; Markus Schmid; FT Modulates Genome-Wide DNA-Binding of the bZIP Transcription Factor FD. Plant Physiology 2019, 180, 367-380, 10.1104/pp.18.01505.
- Ayako Yamaguchi; Miin-Feng Wu; Li Yang; Gang Wu; R. Scott Poethig; Doris Wagner; The MicroRNA-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Developmental Cell 2009, 17, 268-278, 10.1016/j.devcel.2009.06.007.
- Jae-Hoon Jung; Hyo-Jun Lee; Jae Yong Ryu; Chung-Mo Park; SPL3/4/5 Integrate Developmental Aging and Photoperiodic Signals into the FT-FD Module in Arabidopsis Flowering. Molecular Plant 2016, 9, 1647-1659, 10.1016/j.molp.2016.10.014.
- S. Vinod Kumar; Doris Lucyshyn; Katja E. Jaeger; Enriqueta Alós; Elizabeth Alvey; Nicholas P. Harberd; Philip A. Wigge; Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242-245, 10.1038/nature10928.
- Vinicius Costa Galvāo; Anne-Sophie Fiorucci; Martine Trevisan; Jose Manuel Franco-Zorrilla; Anupama Goyal; Emanuel Schmid-Siegert; Roberto Solano; Christian Fankhauser; PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nature Communications 2019, 10, 1-10, 10.1038/s41467-019-11882-7.
 Encyclopedia
Encyclopedia



