Cardiovascular disease (CVD) is the leading cause of morbidity and mortality in adults in developed countries. CVD encompass many diseased states including hypertension, coronary artery disease and atherosclerosis. Studies in animal models and human studies have elucidated the contribution of many genetic factors, including non-coding RNAs. Non-coding RNAs are RNAs not translated into protein, involved in gene expression regulation post-transcriptionally and implicated in CVD. These include long non-coding RNAs. Of these, circular RNAs (circRNAs) and micro RNAs. CircRNAs are created by the back-splicing of pre-messenger RNA and have been underexplored as contributors to CVD. These circRNAs may also act as biomarkers of human disease as they can be extracted from whole blood, plasma, saliva and seminal fluid. CircRNAs have recently been implicated in various disease processes, including hypertension and other cardiovascular disease. This review article will explore the promising and emerging roles of circRNAs as potential biomarkers and therapeutic targets in CVD, in particular hypertension.
- long non-coding RNA
- circular RNA
- micro RNA
- hypertension
- coronary artery disease
- atherosclerosis
- cardiovascular disease
- heart disease
CircRNAs are abundant, underexplored ncRNAs. Recent studies revealed that large numbers of circRNAs are endogenous, highly conserved and stable in mammalian cells and prevalent in disease states (Figure 2) [7]. Although mRNA and circRNAs both originate from precursor-mRNAs, they are formed differently, giving them unique characteristics. mRNAs are formed by RNA splicing where introns are removed, and certain exons are included or excluded to create alternative coding mature mRNAs. This process creates linear mRNAs with exposed poly(A) tails. This characteristic leaves them prone to degradation by RNases [55]. Meanwhile, circRNAs are formed by back-splicing, promoting the circularization process where exons and/or introns converge onto each other, potentially protecting them from degradation and conferring a half-life of approximately 48 h, around five times more stable than linear mRNAs (Figure 1) [56].
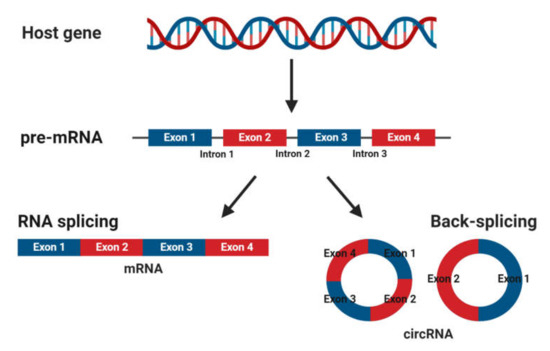
Figure 1. Biogenesis of mature messenger RNA (mRNA) and circular RNA (circRNA) from precursor mRNA (pre-mRNA). Pre-mRNA undergoes RNA splicing to form a mature mRNA or back-splicing to form circRNAs.
The definitive function of circRNAs still remains unclear. It has been proposed that circRNAs regulate the expression of linear mRNA transcripts both directly, via the competition with the splicing machinery [57]; and indirectly, acting as sponges to miRNAs due to the presence of multiple binding sites, allowing them to interact with and sequester cellular miRNAs preventing the performance of their roles on post-transcriptional regulation (Figure 2) [58,59].
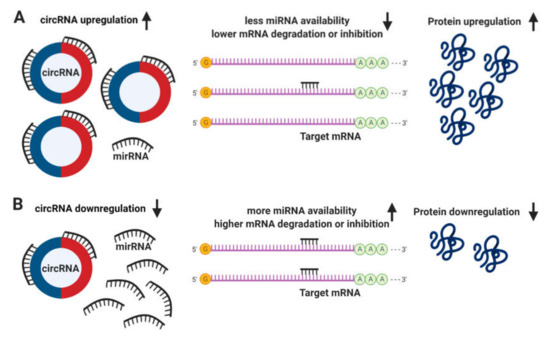
Figure 2. Circular RNA (circRNA) functions. (A) The upregulation of circRNAs leads to a decrease in microRNA (miRNA) availability, messenger RNA (mRNA) degradation or inhibition, resulting in an increase of protein translation. (B) Conversely, the downregulation of circRNAs leads to more miRNA available and higher levels of mRNA degradation or inhibition, leading to a downregulation in protein translation.
Importantly, circRNAs make up 1% of total RNA being expressed widely in various cell types and may have a regulatory function in human disease, with a pivotal role in the initiation and progression of various types of biological processes [58], potentially acting as a biomarker for the discovery and investigation on the progression of disease. However promising, research to identify and characterize circRNAs has mostly been performed using bioinformatics and in silico approaches and a limited number of studies have investigated their function in situ or in vivo to establish their involvement in disease.
The properties described above and promising research in the field of cancer genomics using circRNAs as biomarkers is encouraging and should be explored and translated into cardiovascular genomics research. This is extremely relevant, as conventional methods for controlling risk factors and initiating early treatment in CVD intervention have often led to poor prognosis. Current biomarkers usually detect the disease at later stages of development, increasing the need for the discovery of new biomarkers for prevention or at early onset of disease. This further highlights the benefits of using circRNAs as potential biomarkers in CVD (Table 1).
Emerging evidence of circular RNAs in cardiovascular disorders has demonstrated differential expression in both healthy and diseased hearts [60–63]. However, the relevance of circular RNAs to the cardiovascular system remains poorly characterised, and an improvement in understanding of circRNA involvement in CVD will form a basis for the development of these RNAs as biomarkers for discovery, prediction and therapeutic agents. Importantly, the combination of genetic sequencing and bioinformatics discovery has enabled the identification of many novel circRNAs.
Table 1. Summary of circRNAs implicated in cardiovascular diseases, including species and sample types studied and differential expression direction.
|
Disease |
Circular RNA |
Species |
Sample |
Expression |
References |
|
Hypertension |
hsa_circ_0005870 |
Human |
Blood |
Down |
[64] |
|
hsa_circ_0037909 |
Human |
Blood |
Up |
[65] |
|
|
hsa_circ_0037911 |
Human |
Blood |
Up |
[66] |
|
|
hsa_circ_0126991 |
Human |
Blood |
Up |
[67] |
|
|
hsa_circ_0014243 |
Human |
Blood |
Up |
[68] |
|
|
Myocardial infarction |
mmu_circ_0001878 |
Mouse |
Cardiomyocytes |
Up |
[69] |
|
circTtc3 |
Rats |
Cardiomyocytes |
Up |
[70] |
|
|
MICRA |
Human |
Blood |
Down |
[71] |
|
|
circNfix |
Mouse |
Cardiomyocytes |
Up |
[72] |
|
|
circFndc3b |
Mouse Human |
Cardiomyocytes |
Down |
[73] |
|
|
Atherosclerosis |
circRNA-0044073 |
Human |
Blood |
Up |
[74] |
|
ocu-ciR-novel-18038 |
Rabbit |
Blood |
Down |
[75] |
|
|
ocu-ciR-novel-18298 |
Rabbit |
Blood |
Up |
||
|
ocu-ciR-novel-15993 |
Rabbit |
Blood |
Up |
||
|
ocu-ciR-novel-17934 |
Rabbit |
Blood |
Down |
||
|
ocu-ciR-novel-17879 |
Rabbit |
Blood |
Up |
||
|
ocu-ciR-novel-18036 |
Rabbit |
Blood |
Up |
||
|
ocu-ciR-novel-14389 |
Rabbit |
Blood |
Up |
||
|
circANRIL |
Human |
Blood |
Up |
[76] |
|
|
circCHFR |
Human |
VSMC |
Up |
[77] |
|
|
circRNA-0003575 |
Mouse |
Endothelial cells |
Up |
[78] |
|
|
Coronary artery disease |
hsa_circ_0001879 |
Human |
Blood |
Up |
[79] |
|
hsa_circ_0004104 |
Human |
Blood |
Up |
||
|
hsa_circ_0124644 |
Human |
Blood |
Up |
[80] |
|
|
hsa_circ_0098964 |
Human |
Blood |
Up |
||
|
hsa_circ_0030769 |
Human |
Blood |
Up |
[81] |
|
|
hsa_circ_0079828 |
Human |
Blood |
Up |
||
|
hsa_circ_15486-161 |
Human |
Blood |
Up |
||
|
hsa_circ_0122274 |
Human |
Blood |
Up |
||
|
hsa_circ_16316-13 |
Human |
Blood |
Up |
||
|
hsa_circ_0140538 |
Human |
Blood |
Up |
||
|
Cardiomyopathy |
mmu_circ_0000254 |
Mouse |
Cardiomyocytes |
Down |
[82] |
|
hsa_circ_0076631 |
Human |
Cardiomyocytes and Serum |
Up |
[83] |
|
|
circRNA_000203 |
Mouse |
Ventricular Cardiomyocytes |
Up |
[84] |
|
|
circSlc8a1 |
Mouse |
Cardiomyocytes |
Up |
[85] |
Legend: ciR, circular; circ, circular; circANRIL, circular antisense non-coding RNA in the INK4 locus; circCHFR, circular checkpoint with fork-head associated and ring finger; circFndc3b, circular fibronectin type III domain containing 3B; circNfix, circular nuclear factor I X; circSlc8a1, circular solute carrier family 8 member a1; circTtc3, circular tetratricopeptide repeat domain 3; hsa, Homo sapiens; MICRA, myocardial infarction-associated circular RNA; mmu, Mus musculus; ocu, Oryctolagus cuniculus; VSMC, vascular smooth muscle cells.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21103666
 Encyclopedia
Encyclopedia



