Please note this is a comparison between Version 1 by Honghong Wu and Version 2 by Lily Guo.
Salinity is an issue impairing crop production across the globe. Under salinity stress, besides the osmotic stress and Na+ toxicity, ROS (reactive oxygen species) overaccumulation is a secondary stress which further impairs plant performance. Chloroplasts, mitochondria, the apoplast, and peroxisomes are the main ROS generation sites in salt-stressed plants.
- ROS Scavenging
- antioxidant enzymes
- non-enzymatic scavenging system
1. Enzymatic Scavenging System
Protection of chloroplasts from ROS accumulation is very important for stress tolerance in plants. Superoxide dismutase (SOD), ascorbate peroxidase (APX), and glutathione reductase (GR) constitute an efficient ascorbate–glutathione cycle (ASA–GSH, Figure 13), which is a part of the enzymatic antioxidant system of plants [1][2][3][4][5][75,76,77,78,79]. In chloroplasts, a major ROS-scavenging pathway in plants is the water–water cycle (WWC). WWC can protect enzymes in chloroplasts and PSI from ROS and dissipate excess light energy in plant cells [6][80].
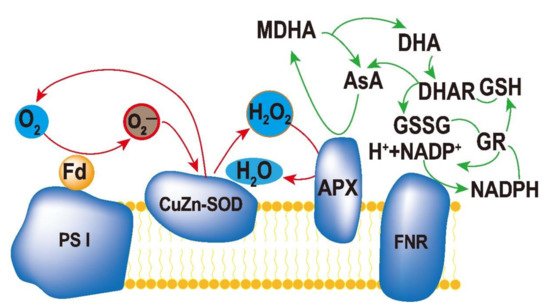
Figure 13.
Ascorbate–glutathione cycle in plant cells. The reduced form of ascorbate (Asc) is oxidized to monodehydroascorbate (MDHA), which can either be reduced by monodehydroascorbate reductase (MDHAR) to Asc, or react to dehydroascorbate (DHA). DHA can be reduced by dehydroascorbate reductase (DHAR) to Asc. The reduced form of glutathione (GSH) is oxidized to glutathione disulfide (GSSG), which is reduced by glutathione reductase (GR) to GSH. During the reduction of MDHA and GSSG, the electron acceptor NADP is regenerated.
-
(1) 2H2O → 4[e−] + 4H+ + O2 (The photolysis of H2O in PSII);
-
(2) 2O2 + 2[e−] → 2 O2− (Photoreduction of O2 in PSI);
-
(3) 2O2− + 2H+ → H2O2 + O2 (SOD catalyzes O2− disproportionation);
-
(4) H2O2 + 2AsA → 2H2O + 2MDA (APX catalyzes ASA reduction H2O2);
-
(5) 2MDA + 2[e−] + 2H+ → 2AsA.
-
Total: 2H2O + O2− → O2 + 2H2O.
A previous study indicated that optimum exogenous selenium enhanced the activity of POD and APX, which ultimately protected chloroplasts from auto-pepsia and resulted in a higher photosynthesis rate in maize crop grown under salt stress [7][81]. Transcriptomic and metabolomic analyses revealed that genes encoding LEA (late-embryogenesis-abundant) proteins are up-regulated under salt stress. It is evident that LEA proteins could enhance the antioxidant enzyme activities [8][82]. It is evident that halophytes have a stronger enzymatic antioxidant system (CAT and SOD), and this capability has been used as a selection criterion for salt-tolerant varieties [9][10][11][83,84,85]. A previous study reported that SOD converted salt-induced ROS into H2O2, and APX maintained H2O2 at an optimum level as a signal molecule [12][86]. The study also revealed the importance of PaSOD (superoxide dismutase) and RaAPX (ascorbate peroxidase) gene in plant salt tolerance, which could enhance the biosynthesis of compatible solutes and lignin. Overexpression of mitochondria-located protein 4-hydroxybenzoate polyprenyl diphosphate transferase (PPT) increased the ubiquinone content and enhanced the plant’s salt-tolerance. Glutathione peroxidase (GPX) is an important component of the ASA–GSH cycle. The ASC (ascorbate)/GSH cycle is also an important antioxidant system in plants. A study on the glycophyte Brassica juncea and the halophyte Sesuvium portulacastrum indicated that the ASC/DHA (dehydroascorbate) and GSH/GSSG (glutathione disulfide) ratios were maintained in B. juncea but increased in S. portulacastrum [11][85]. By using OsGPX3-RNAi silenced plants, researchers found that GPX had an important role in plant CO2 assimilation, biomass accumulation, and PS II protection under salt stress [13][87]. Interestingly, OsGPX3-RNAi silenced plants displayed no observable difference with untransformed plants in ROS accumulation, suggesting a complex role for GPX in plant response to salt tolerance [14][88]. In addition, the gene related to alternative oxidase (AOX) located in the inner membrane of mitochondria has been reported, and is believed to be involved in the response to abiotic stress [15][89].
2. Non-Enzymatic Scavenging System
Several highly toxic ROS, such as 1O2 and •OH, cannot be scavenged by enzymatic antioxidant systems. Thus, plants depend solely on non-enzymatic means to scavenge those ROS. Generally, ROS lead to lipid peroxidation [16][17][90,91] and an increase in monodehydroascorbate (MDA) content [18][92]. Overexpression of IbWRKY2 induced MDA accumulation in plant cells, coupled with ROS-scavenging genes up-regulation, resulting in improved salt tolerance [19][18]. Carotenoids and α-tocopherol, as components of non-enzymatic antioxidant systems, can also scavenge ROS. For example, carotenoids are distributed in the thylakoid pigment—protein complexes in close proximity to chlorophylls near the potential sites of 1O2 formation. Furthermore, melatonin is considered a small molecule that may scavenge ROS, and the “manager” in plants under salinity stress. For example, a previous study found that the gene related to the melatonin synthesis enzyme N-acetylserotonin O-methyltransferase (MzASMT9), which is localized in the chloroplast, is upregulated by salt stress [20][93]. Additionally, MTs (metallothioneins) interact with AtVDAC3 (mitochondrial membrane voltage-dependent anion channels) and regulate ROS homeostasis [21][45]. Flavodoxin is known as an electron transfer shuttle, and can drive reducing equivalents away from oxygen, preventing ROS generation [22][94]. However, interestingly, compared with the enhanced tolerance to drought, excess irradiation, temperature stress, and iron starvation, overexpression of the plastid-targeted cyanobacterial flavodoxin in tobacco plants failed to result in increased salt resistance [23][95].
 Encyclopedia
Encyclopedia

