The gene coding for the telomerase reverse transcriptase (TERT) is essential for the maintenance of telomeres. Previously we described the presence of three TERT paralogs in the allotetraploid plant Nicotiana tabacum, while a single TERT copy was identified in the paleopolyploid model plant Arabidopsis thaliana. Here we examine the presence, origin and functional status of TERT variants in allotetraploid Nicotiana species of diverse evolutionary ages and their parental genome donors, as well as in other diploid and polyploid plant species. A combination of experimental and in silico bottom-up analyses of TERT gene copies in Nicotiana polyploids revealed various patterns of retention or loss of parental TERT variants and divergence in their functions. RT–qPCR results confirmed the expression of all the identified TERT variants. In representative plant and green algal genomes, our synteny analyses show that their TERT genes were located in a conserved locus that became advantageous after the divergence of eudicots, and the gene was later translocated in several plant groups. In various diploid and polyploid species, translocation of TERT became fixed in target loci that show ancient synapomorphy.
1. Introduction
Flowering plants (angiosperms) are important for the existence of many terrestrial organisms, including humans, and a long history of plant breeding has taught us that polyploidization can be advantageous in terms of quantitative traits of crops. Gains and losses of paralogs, their neofunctionalization and sub-functionalization, have all been associated with the generation of duplicate gene copies, e.g., by whole-genome duplications (WGDs) and further rounds of genome duplication/reduction, resulting in genetic diversity upon which the fittest combinations thrived in a competitive environment
[1,2,3,4][1][2][3][4]. An ancient WGD has been reconstructed at the base of seed plants, another at the base of angiosperms
[5,6,7][5][6][7] and numerous additional, subsequent WGD events were associated with the divergence of many angiosperm lineages
[3]. Polyploidy is usually associated with many genetic and epigenetic changes, including chromosomal rearrangements, expansions of transposable elements and changes in gene expression
[8,9][8][9]. At the gene level, polyploids can tolerate the presence of paralogs or eliminate a copy of the spare gene. Thus, evolutionary forces result in an equilibrium defined by gene dosage
[10]. Studies of model plants have mostly focused on genes important for crop production; however, genes that are critical for genome stability are extremely important for understanding repeated polyploidization events during natural selection, and these remain underexplored.
Telomerase reverse transcriptase (TERT) is involved in the maintenance of telomeres, nucleoprotein structures that are essential for genome stability
[11,12,13][11][12][13]. Telomerase adds telomere repeats to the ends of eukaryotic chromosomes, thereby elongating telomeres and compensating for their shortening due to incomplete end-replication. When telomerase is not active, telomeres become shortened, and their function in the protection of chromosomes is disrupted. The extreme evolutionary success of telomerase-based mechanisms of telomere maintenance is illustrated by current findings in plants (reviewed in
[14]). Even among apparent exceptions in telomere sequences, in plant genera
Allium (Asparagales) and
Cestrum (Solanales)
[15,16[15][16][17][18],
17,18], recent research has revealed that novel, unusual telomere DNA sequences are synthesized by telomerase
[16,18,19][16][18][19] and not by alternative mechanisms as had been suggested previously (reviewed in
[20]). Moreover, we recently demonstrated that changes in the template region of the telomerase RNA subunit directed the observed evolutionary transitions in telomere DNA sequences
[14,21,22][14][21][22]. In contrast to the RNA subunit, the protein subunit TERT is evolutionary well conserved and possesses a central reverse transcriptase domain essential for its catalytic function
[23,24][23][24]. Plant TERTs are structurally similar to human, ciliate or yeast TERTs with a telomerase-specific T motif
[25,26,27,28,29][25][26][27][28][29]. The gene encoding TERT is usually expressed at low mRNA levels even in telomerase-positive tissues and is maintained as a single copy gene in most eukaryotic genomes. However, the natural allotetraploid
Nicotiana tabacum possesses three sequence variants of the
TERT gene
[30]. Various allopolyploidization events among closely and distantly related diploid parental species () in
Nicotiana make the genus an ideal experimental model system to study the long-term evolution of
TERT following natural gene duplication. The increasing number of publicly available assembled plant genomes enables the exploration of
TERT genomic loci, gene copy numbers and gene synteny in diverse plant species for comparisons with the data from
Nicotiana polyploids and the diploid species most closely related to their progenitors (hereafter called progenitor diploids). The
Nicotiana genus
[31,32,33,34,35][31][32][33][34][35] comprises relatively young polyploids (i)
N. tabacum (section Nicotianae),
N. rustica (sect. Rusticae),
N. arentsii (sect. Undulatae) that formed approx. 0.4–0.6 million years ago, (ii)
N. clevelandii and
N. quadrivalvis (ca. 1.5 million years ago, sect. Polydicliae), (iii) four species from the 4–5 million years old section Repandae (
N. nudicaulis,
N. repanda,
N. nesophila and
N. stocktonii), and (iv) ~35 species including the model
N. benthamiana from the oldest section Suaveolentes formed about 6 million years ago
[31]. Among these species, members of sections Suaveolentes and Repandae are of interest because, with
N. tabacum, they share an ancient genome donor,
N. sylvestris, and these speciation events happened at different times. In
N. tabacum, two
TERT variants originated from the maternal
N. sylvestris genome (
TERT_Cs,
TERT_D) and one from the
N. tomentosiformis paternal genome (
TERT_Ct). Variants
TERT_Cs and
TERT_Ct code for a full-length functional protein, while the
TERT_D variant is truncated and contains several indels resulting in premature stop codons, suggesting that it is a pseudogene
[30]. All three variants are nevertheless transcribed and show distinct, tissue-dependent levels of mRNA transcripts, indicating a sub-functionalization of
TERT variants
[30,36][30][36].
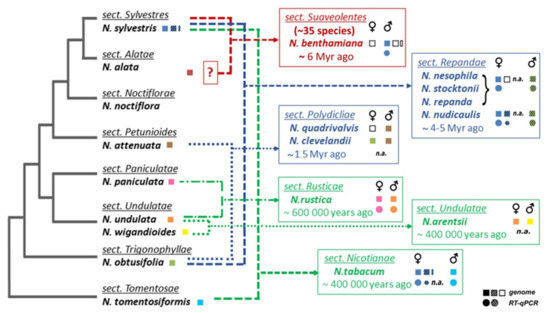
Figure 1. Overview of experimental results and illustration of phylogenetic relationships of
Nicotiana species used in this study. Phylogeny and the proposed origin of polyploids were adapted from
[31,32][31][32]. An uncertain parental genome donor for section Suaveolentes is indicated by a question mark. Summary of experimental and in silico results (squares, analyses of genomic DNA; circles, expression of
TERT variants investigated by RT–qPCR) is shown in boxes of Nicotiana sections, the origin of
TERT variant in polyploids is depicted by color of respective parental diploids, and variants that were not identified are depicted with open squares.
Nicotiana accessions used in the experimental analyses are listed in
Table S1, genomic assemblies and genomic/transcriptomic SRA data used for in silico analyses are listed in Material and Methods. For the purposes of this paper, we refer to a
TERT copy that does not code for a catalytically active protein as a putative pseudogene (dashed symbols) in contrast to a functional
TERT gene copy (open symbols), n.a. not analyzed.
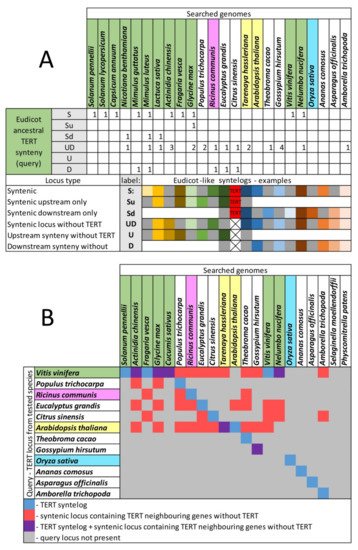
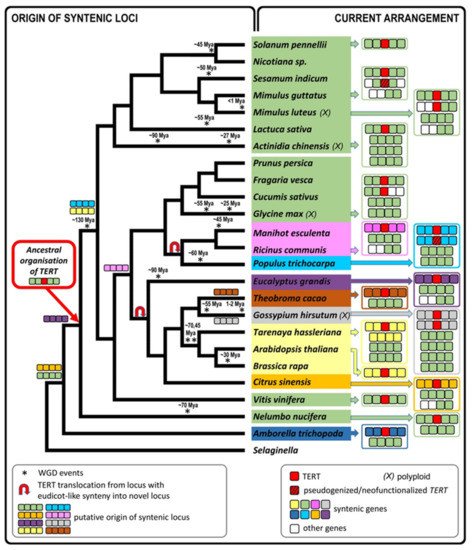
Based on previously described TERT variants in N. tabacum, we explored the fate of TERT paralogs in other Nicotiana polyploids to determine whether both parental TERT genes are conserved in allotetraploid genomes, whether they are transcribed, present in syntenic, collinear arrays with their progenitor diploids, and whether any relationship exists between telomere lengths in polyploids and their progenitor diploids. Of particular interest in this study was to clarify the origin of the presumed pseudogene variant TERT_D in N. sylvestris, a diploid genome donor of N. tabacum, as well as of even older species from sections Repandae and Suaveolentes. In addition, we investigated in silico whether diploid and polyploid plants outside of the family Solanaceae sustained TERT paralogs/pseudogenes in their genomes, and we explored syntenic relationships of genes adjacent to TERT to interpret the evolutionary success of TERT copies after translocation.
2. Number of TERT Variants in Nicotiana Polyploids as a Case Study
At the beginning of this project, there was limited genomic sequence data available for the majority of
Nicotiana allopolyploids and their parents. To characterize experimentally the number, identity and origin of
TERT copies in genomes of polyploid
Nicotiana species and representatives of their diploid progenitors, we employed several primer combinations derived from conserved
TERT regions of the evolutionarily distant relatives
N. sylvestris and
N. tomentosiformis (), designed originally for amplification of
N. tabacum TERT variants
[30,36][30][36]. These PCR primers (A,
Table S2) amplify
TERT regions nonspecifically, i.e., all variants are produced in a single PCR. Sequencing of PCR products then identifies single nucleotide polymorphisms (SNPs) and/or indels evidencing the presence of multiple
TERT variants. Primer positions were with respect to
Nicotiana TERT gene structure with 13 exons (A), which differed from the prevalent 12-exon structure of plant
TERTs
[23]. As expected, a successful amplification was achieved mostly using primers derived from the more conserved sequences at the 3′ end of
TERT genes (
Table S3). As the first screening experiment, we applied this approach to six diploid
Nicotiana species investigated as representatives of parental genome donors, including
N. sylvestris as a control, and to nine polyploid
Nicotiana species (). Among parental diploids, we detected one
TERT variant in
N. alata,
N. attenuata,
N. undulata,
N. wigandoides,
N. paniculata and
N. obtusifolia (
Supplementary A1), and two
TERT variants (
TERT_C and
TERT_D) in
N. sylvestris [30]. In the case of
N. attenuata and
N. obtusifolia, species representing parents of polyploid sections Polydicliae and Repandae, we further confirmed our results by in silico analysis using genome assemblies (GenBank accessions: GCA_001879085.1 and GCA_002018475.1, respectively). To complete the set of representative parental species, we assembled available transcriptomic SRA data of
N. noctiflora (GenBank accession: SRR2106514) and identified one
TERT variant. In conclusion, our results show the presence of more than one
TERT variant in diploid
N. sylvestris [30], an exception among parental species of
Nicotiana polyploids.
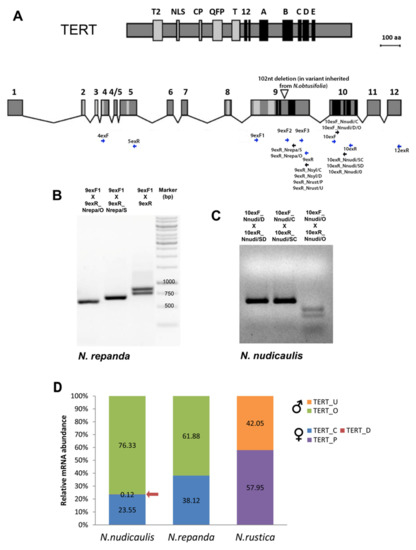
Figure 2. Experimental identification of TERT variants and analysis of gene expression in
Nicotiana polyploids. (
A) Conserved telomerase specific motifs (T2, NLS, CP, QFP, T) and reverse transcriptase motifs (1, 2, A–E) are highlighted in protein and mRNA of
Nicotiana TERT (modified from
[30]). Positions of primers used for screening experiments (blue arrows) and
TERT-variant-specific primers (black arrows) are indicated at corresponding
TERT mRNA regions (primers are listed in
Table S2). The triangle within exon 9 shows the position of a 102 nt long deletion that was identified in
N. repanda,
N. nesophila and
N. stocktonii and represents a specific
TERT-variant of
N. obtusifolia origin. (
B,
C) Validation of primer specificity for
TERT variants in
N. repanda (
B) and
N. nudicaulis (
C). PCR products amplified with primers 9exF1 and 9exR1 show two bands corresponding to
TERT_O and
TERT_Cs variants that differ by a 102 bp long deletion. Specific amplification of
TERT_O and
TERT_Cs variants was demonstrated using the 9exF1 primer in combination with variant-specific reverse primers 9exR_Nrepa/O and 9exR_Nrepa/S, respectively. (
C) For validation of qPCR primers and to distinguish three
TERT variants in
N. nudicaulis, the PCR products amplified with indicated qPCR primer combinations were digested with
MseI. A specific cut of the
TERT_O variant that possesses the restriction site for
MseI within the amplified region confirmed the specificity of amplified
TERT-variants. (
D) Relative mRNA levels of specific
TERT variants were determined by RT–qPCR in
N. nudicaulis,
N. repanda and
N. rustica. Relative mRNA abundance of particular parental
TERT variants (in %) was calculated by the delta Ct method
[37]. Ct values were normalized using the reaction efficiency calculated from a standard curve analysis (
Table S3).
The same experimental approach applied to representative
Nicotiana polyploids detected variant-specific SNPs and/or indels, demonstrating the presence of two
TERT variants in 5 of 9 polyploid species investigated (
N. arentsii,
N. rustica,
N. repanda,
N. nesophila,
N. stocktonii) and three variants were identified in
N. nudicaulis (summarized in , , see below for details). While PCR products obtained from
N. clevelandii, N. quadrivalvis and
N. benthamiana genomic DNA revealed the presence of a single copy of the
TERT gene, our search for
TERT variants in raw transcriptomic data from
N. clevelandii showed the occurrence of two gene variants. To avoid possible errors in comparison of experimental and in silico data that could be caused, e.g., by possible incorrect mapping of
TERT reads to the raw genome/transcriptome data, assembly version or allele sequence, we analyzed in detail individual SNPs in sequences from each polyploid species and its progenitor diploids (see
Supplemental Text S1,
Figure S1,
Table S4). Results deduced from sequence similarity (in %, ) and individual SNPs (
Table S4) were in agreement in all cases analyzed.
Table 1. Origin of telomerase reverse transcriptase (TERT) variants in polyploid Nicotiana species determined by sequence similarity with representative progenitor diploids.
| Allopolyploids |
GeneBank
Accessions |
Sequence Similarity [%] |
Analyzed Region | 1 |
| Maternal Parent |
Paternal Parent |
| SUAVEOLENTES |
|
N. alata |
N. noctiflora2 |
N. syl. C var. |
N. syl. D var. |
|
TERT copies after young polyploidization events is obvious ().
Table 2. Number of TERT gene copies in Nicotiana species determined in silico.
| Species/Genome Dataset Accession |
Total No. of TERT Reads |
Expected Genome Coverage (Depth) |
No. of Detected TERT Variants |
Read Counts Corresponding to Known TERT Variants |
Ratio of TERT Variants in Genome |
| N. benthamiana |
| NbS000104 |
96.3 |
n.a. |
97.5 |
n.a. |
exon 4 to 5 |
| 27g0116.1 |
n.a. |
96.1 |
97.6 |
93.4 |
exons 10, 11, 12 |
| REPANDAE |
|
N. syl. C var. |
N. syl. D var. |
N. obtusifolia |
|
| N. repanda |
MG242402 | 1 |
95.9 |
91.6 |
97.4 |
exon 9 |
| MG242403 | 1 |
97.9 |
92.4 |
96.4 |
exon 9 |
| N. stocktonii |
MG242407 | 1 |
95.6 |
91.7 |
97.6 |
exon 9 |
| MG242408 | 1 |
98.6 |
93.1 |
97.0 |
exon 9 |
| N. nesophila |
MG242405 | 1 |
95.2 |
91.6 |
97.0 |
exon 9 |
| MG242406 | 1 |
98.5 |
92.9 |
96.9 |
exon 9 |
| N. nudicaulis |
MG242409 | 1 |
98.6 |
94.3 |
94.4 |
exon 10 to 12 |
| MG545647 | 1 |
92.8 |
94.8 |
91.6 |
exon 10 to 12 |
| MG242410 | 1 |
94.2 |
93.3 |
96.3 |
exon 10 to 12 |
| POLYDICLIAE |
|
N. obtusifolia |
N. attenuata |
|
| |
MG242422 | 1 |
94.3 |
99.3 |
exon 4 to 5 |
| N. clevelandii |
var1 | 2 |
97.3 |
98.9 |
exon 9 | 2 |
| |
var2 | 2 |
99.2 |
97.3 |
exon 9 | 2 |
| N. quadrivalvis |
MG242423 | 1 |
94.9 |
98.6 |
exon 4 to 5 |
| ARENTSII |
|
N. undulata |
N. wigandiodes |
|
| N. arentsii |
MG242418 | 1 |
99.5 |
98.4 |
exon 9 |
| MG242419 | 1 |
98.8 |
99.8 |
exon 9 |
| RUSTICA |
|
N. paniculata |
N. undulata |
|
| N. rustica |
MG242413 | 1 |
100.0 |
98.2 |
exon 9 |
| MG242414 | 1 |
98.2 |
99.8 |
exon 9 |
 Encyclopedia
Encyclopedia

