1000/1000
Hot
Most Recent

Viruses are dependent on host factors at all parts of the infection cycle, such as translation, genome replication, encapsidation, and cell-to-cell and systemic movement. RNA viruses replicate their genome in compartments associated with the endoplasmic reticulum, chloroplasts, and mitochondria or peroxisome membranes. In contrast, DNA viruses replicate in the nucleus. Viral infection causes changes in plant gene expression and in the subcellular localization of some host proteins. These changes may support or inhibit virus accumulation and spread. Here, we review host proteins that change their subcellular localization in the presence of a plant virus. The most frequent change is the movement of host cytoplasmic proteins into the sites of virus replication through interactions with viral proteins, and the protein contributes to essential viral processes. In contrast, only a small number of studies document changes in the subcellular localization of proteins with antiviral activity. Understanding the changes in the subcellular localization of host proteins during plant virus infection provides novel insights into the mechanisms of plant–virus interactions and may help the identification of targets for designing genetic resistance to plant viruses.
The most abundant plant viruses have a genome that is a positive single-strand RNA (Group IV) or a negative single-strand RNA (Group V). Single-strand RNA viruses replicate in compartments or vesicles bound to membranes in the cytoplasm or in subcellular organelles [1]. Plant-infecting DNA viruses, on the other hand, are less numerous. Single-strand DNA (Group II) and reverse-transcribing DNA (Group VII) viruses replicate by forming a minichromosome in the nucleus [2].
Viral RNA is translated into proteins using the cellular machinery. Viral nucleic acids and proteins execute their functions in cooperation with host proteins, RNAs, or other factors such as membranes or lipids [3][4]. These components condition susceptibility, and their absence reduces virus accumulation or movement, and may turn a host into a nonhost. These factors encode loss-of-susceptibility genes, also named susceptibility genes [3][5]. Because the presence and activity of these host components are essential for the virus, the terms cellular factors with proviral activity or proviral host factors are often used in publications [3][6].
The establishment of a viral infection is genetically determined at two sequential phases. Initially, the absence of susceptibility genes results in the lack of infection or reduced virus replication and/or movement [3]. When a plant has the susceptibility genes needed for the initiation of infection, in a second phase, virus accumulation, spread, and disease severity are determined by the balance between plant defense and viral suppression of defense responses [7].
Viral infection induces changes in host gene expression [8][9] resulting in the upregulation of susceptibility genes [10] and activation or downregulation of antiviral defense responses [11][12], and may also lead to up- or downregulation of genes that have no effect on the virus [8][9]. Upregulation of antiviral genes indicates the activation of defense responses by multiple mechanisms including autophagy, RNA decay, or gene silencing [7][13][14]. Antiviral defense is mediated by host factors that target viral proteins or nucleic acids and antagonize key parts of virus replication and/or movement, reducing virus accumulation or limiting the spread of infection within the plant [7][15]. However, to protect themselves, viruses may downregulate expression, suppress activity, or induce degradation of antiviral defense components [16]. The molecular mechanisms and significance of changes in host gene expression during viral infection are still poorly understood.
Viruses divert host proteins from their natural roles to execute essential viral processes such as translation, virus replication, or movement [5][17][18]. Changes in activity are often associated with a change in the subcellular localization of the host protein. A protein is considered to relocalize when, in the presence of a virus, a fraction of the total protein accumulates in a new place in the cell. These changes have been detected and characterized by a combination of approaches such as yeast two hybrid, subcellular fractionation, bimolecular fluorescence complementation, immunofluorescence confocal microscopy, or co-precipitation [19][20][21].
Plant viruses for which at least one host protein has been reported to change subcellular localization were grouped based on their genome organization. The site of genome replication, name of the replication protein, and movement form were compiled and used as a guide to interpret interaction with and recruitment of host proteins (Table 1). We classified the host proteins based on their natural subcellular localization in the absence of viral infection. Changes in subcellular localization were documented for 55 combinations of host protein and plant virus. After profiling features of these proteins and viruses, several general patterns emerged: (1) 45 of the 55 combinations were identified using model hosts (Arabidopsis thaliana, Nicotiana benthamiana, or Saccharomyces cerevisiae, Figure 1A); (2) the majority (48) were identified using model positive-strand RNA viruses (Figure 1C), particularly brome mosaic virus (BMV), tomato bushy stunt virus (TBSV), and turnip mosaic virus (TuMV) (Figure 1D); (3) in 46 of the 55 combinations, the host protein is beneficial to the virus; (4) host proteins with antiviral roles were less abundant (nine) (Figure 1B); and (5) the most frequent group (30 out of 55) was host proteins that moved from the cytoplasm to the sites of virus replication (Figure 2A and Table 2) through interactions with viral proteins (Table 2). These patterns are heavily influenced by the combination of experimental hosts and viruses used as model systems.
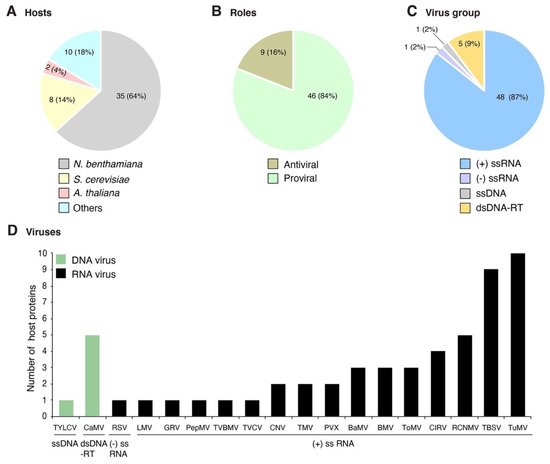
Figure 1. Profile of host proteins that change their subcellular localization during plant virus infection as reported in the literature. Fifty-five combinations of host protein-plant virus were documented in publications. (A) Number and proportion of proteins by host species. (B) Number and proportion of host proteins with antiviral role or beneficial to the virus. (C) Number and proportion of host proteins by virus group. (D) Number of host proteins by virus species. Viruses are grouped based on their genome organization.
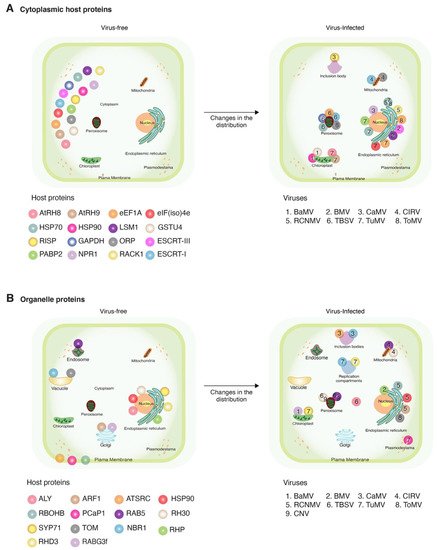
Figure 2. Schematic representation of changes in subcellular localization after viral infection. Representative host proteins and plant viruses that induce relocation in more than two proteins are illustrated. Host proteins are color-coded with spheres. Viruses are indicated by numbers. (A) Changes in host cytoplasmic proteins and (B) changes in host proteins naturally localized to organelles, and their movement in the presence of a virus.