1000/1000
Hot
Most Recent

Colorectal Cancer (CRC) is one of the most common gastrointestinal malignancies which has quite a high mortality rate. Despite the advances made in CRC treatment, effective therapy is still quite challenging, particularly due to resistance arising throughout the treatment regimen. Several studies have been carried out to identify CRC chemoresistance mechanisms, with research showing different signalling pathways, certain ATP binding cassette (ABC) transporters and epithelial mesenchymal transition (EMT), among others to be responsible for the failure of CRC chemotherapies. In the last decade, it has become increasingly evident that certain non-coding RNA (ncRNA) families are involved in chemoresistance. Research investigations have demonstrated that dysregulation of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) contribute towards promoting resistance in CRC via different mechanisms. Considering the currently available data on this phenomenon, a better understanding of how these ncRNAs participate in chemoresistance can lead to suitable solutions to overcome this problem in CRC.
RNAs provide both catalytic and informative functions. With the rapid, increased development of high throughput methods and transcriptome sequencing techniques, scientists have gained a better understanding of both the protein-coding and non-coding portions of the mammalian transcriptome. Most of the mammalian genome is actively transcribed with approximately 2% of the genome encoding proteins [1][2][3]. The remainder of the human genome is known to harbour ncRNAs [4]. NcRNAs are involved in controlling gene expression before, after and throughout transcription, thus these RNAs participate in regulating and modulating different stages tumour progression, namely cell proliferation, migration, metastasis and chemoresistance [5]. The advances in the field of ncRNA have been reviewed from different mechanistic perspectives [6][7][8] and biological roles [3][9][10]. Eukaryotic transcription can generate different ncRNA species, which arise from different genomic regions and RNA processing [11]. Furthermore, different parts of the DNA can be transcribed into ncRNAs, for example protein-coding genes, transposon elements or enhancer regions [11][12]. Considering their average size and their regulatory role, ncRNA are split up into two groups: housekeeping ncRNAs and regulatory ncRNAs which are then further subdivided (Figure 1) [11][12][13][14]. Multiple kinds of ncRNAs have been identified and validated within these two groups, together with their vital functions in both pathological and physiological processes. Most of the identified ncRNA have been linked to different cellular functions, such as gene expression, proliferation, cell cycle progression, apoptosis, and various other functions [15]. Housekeeping ncRNAs are small (ranging from 50 to 500 nucleotides), constitutively expressed in all cell types and needed for cell viability [11][12]. Furthermore, this group primarily regulates generic cellular functions [12][13][14][16][17][18]. The regulatory ncRNAs can be further categorized into two groups based on their size: small non-coding RNAs (sncRNAs), which have transcripts not bigger than 200 nt and long noncoding RNA (lncRNAs) which have transcripts greater than 200 nt [11][12] (Figure 1). These act as key regulatory RNA molecules, which function as gene expression regulators at an epigenetic, transcriptional and post transcriptional level [11][16][17][18]. Even though ncRNAs are not present in great abundance and were referred to as ‘junk’ DNA, these still have important roles in transcription, post-transcriptional processing, and translation and are also constitutively expressed and essential for normal function of the cell [16][19][20].
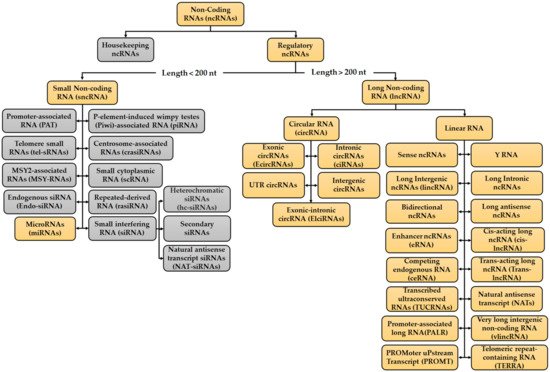
Figure 1. Classification of the Non-coding RNA (ncRNA) families. Two types of ncRNAs have been identified, Housekeeping ncRNAs and Regulatory ncRNAs. Despite the numerous types of ncRNAs identified, the focus in this review will be on the ones highlighted in orange as the ncRNAs to be discussed in the chemoresistance sections fall under these categories. Further work is to be done on the ncRNAs present in the grey boxes in order to see their involvement in CRC chemoresistance. Information for figure retrieved from: [4][11][13][14][21][22][23][24][25][26][27][28][29][30][31][32].
The majority of the currently published/reported studies are on miRNAs and lncRNAs. However, in addition to miRNA and lncRNA, another type of ncRNA, circRNA, has also recently became one of the research focuses. MiRNAs are said to be the most abundant class of small ncRNAs, produced from transcribed hairpin loop structures [33][34]. MiRNAs contribute to regulating gene expression in the nucleus and cytoplasm via different mechanisms and mediate gene silencing at a post-transcriptional level in many human cellular contexts and diseases [11][33]. When carrying out their functions, miRNAs tend to also interact with other types of ncRNAs, mainly with lncRNAs and circRNAs, as these help regulate miRNA stability [33][34][35]. Furthermore, they contribute to differentiation and development (cell growth and survival) in human cells by completely or incompletely binding to the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs), which results in mRNA degradation or post-transcriptional inhibition [36]. In addition, miRNAs also contribute to regulating different biological pathways, including the cell cycle, apoptosis, differentiation, DNA repair, energy metabolism, proliferation, immune response, and stress tolerance [37][38]. LncRNAs are mostly transcribed by RNA polymerase II, but there are some cases in which lncRNAs are generated from the antisense region of upstream promoters [39][40]. They do not have an open reading frame, often harbor a poly-A tail, can be spliced, and are generally found in the cytoplasm and nucleus of a cell [20][39][41][42]. LncRNAs found in the nucleus are functionally involved in gene regulatory procedures, such as promoter specific repression or activation of transcription [7][43][44][45] or epigenetic gene regulation [46][47] and those found in the cytoplasm modulate post-transcriptional gene regulatory processes [48][10][20][49][50][51]. In addition, they modulate protein interactions, stability and affect their cellular localisation, together with controlling signalling cascades and any changes in gene expression [47]. CircRNAs are a category of endogenous ncRNA molecules which are widely expressed in eukaryotic cells and exhibit both location- and step-specificity [52][53]. As their name implies, they are circular (closed loops) and arise from splicing events and are one of the ncRNAs which can be generated from protein coding regions, but can also be synthesised from introns, intergenic regions, untranslated regions or from tRNAs [11][54][55]. This class of ncRNAs are mostly found in the cytoplasm, however a few can reside in the nucleus with most circRNAs having a conserved sequence [54][56]. Just like the other regulatory ncRNAs, circRNAs are involved in different biological processes; (1) interact with RNA binding proteins since circRNAs harbour sites for these proteins which can serve as protein sponges or decoys during gene expression [57], (2) bring about epigenetic alterations [58], (3) act as regulators for transcription or post-transcription and alternative RNA splicing [59][60], (4) translate proteins [53]. Despite their functional roles in cells, their biological roles remain largely speculative. Something common between both lncRNAs and circRNAs is that they can both act as miRNA sponges or competing endogenous RNAs (ceRNAs) to competitively bind to and sequester miRNAs, decreasing their regulatory effect on mRNAs [47][61][62][63]. CeRNAs refers to different ncRNAs (e.g., circRNAs and lncRNAs) which compete (through interactions/crosstalk) for the same pool of miRNAs, thus in turn regulate the activity of miRNAs [64]. miRNA sponge is another term used instead of ceRNAs and refers to those ncRNAs which attract miRNAs for binding and competitively sequester them from the miRNAs natural target/s due to having multiple tandem high-affinity binding sites to that miRNA of interest [65]. For both cases (miRNA sponges or ceRNAs), the miRNAs targeted will reduce its regulatory effect on mRNAs due to miRNAs being negatively regulated by other ncRNAs, thus releasing the inhibition of the mRNA being targeted by the miRNAs so degradation or translation can take place.
The normal function and expression of the different ncRNAs are vital for maintaining physiological conditions. However, if there is any disturbance in the function or abnormal expression of said ncRNAs, these can contribute to the development of pathological events, mainly cancer. Several studies have shown that aberrant expression of miRNAs, lncRNAs and circRNAs may participate in CRC progression via different mechanisms and in turn contribute to cancer cell growth and proliferation, apoptosis, metastasis, angiogenesis, and EMT transition [3][20][33][42][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88]. This dysregulation results in the alteration of several signalling pathways, but due the complex structural characteristics demonstrated by ncRNAs, further structural, functional, and mechanistic characterisations are required to get a better understanding of their roles in cancer development [89]. Furthermore, throughout cancer development these three ncRNAs can act as oncogenes, proto-oncogenes or as tumour suppressors, with some ncRNAs also acting as both oncogenes and tumour suppressors [13][90][91][92][93][94][95][96][97]. Research shows that apart from ncRNAs being dysregulated, they can potentially be used as diagnostic and prognostic markers when collected from tissues, plasma, or serum for specific cancers such as CRC, pancreatic cancer, breast cancer, leukemia, lung cancer and various others [98][99]. Furthermore, due to having tissue specific signatures and due to the expression patterns, they demonstrate in tumours, ncRNAs have shown to be promising for the generation of accurate non-invasive biomarkers for prognosis and diagnosis [89]. Most of the ncRNA families have shown to contribute to not only cancer development but can also give rise to chemoresistance [15][100][101][102][103][104], and the coming sections will describe chemoresistance arising due to these three ncRNAs in relation to CRC.
Drug resistance is the decrease in effectiveness for drugs including chemotherapeutic agents, antibiotics, and antivirals, throughout the treatment of different diseases. A tumour can be intrinsically drug resistant from the start of treatment or can develop resistance throughout the course of said treatment [105][106]. Chemoresistance can be divided into two: single drug resistance and multidrug resistance (MDR). MDR refers to the development of resistance by cancer cells to anticancer drugs having different structures and functions [107]. This phenomenon typically arises due to various mechanisms and involves intrinsic and extrinsic factors, such as drug inactivation, EMT, DNA repair alterations and epigenetics, inhibition of cell death related pathways (apoptosis, senescence, necrosis, autophagy), drug efflux and reduced drug uptake, intracellular signalling pathways, changes in membrane lipids, tumour microenvironment, cancer stem cells (CSCs) and membrane transporters [108][109][110][111]. Different mechanisms have been shown to contribute to chemoresistance in CRC when patients are treated with the currently available CRC treatments, such as 5-FU and its analogues, OXA, Cisplatin or DOX. Given the accumulating literature regarding this field, the mechanisms which have shown to give rise to chemoresistance in CRC will be discussed below.
Transport based cellular mechanisms of drug resistance have been shown to be one of the most understood modes of resistance in CRC [112][106][113][114][115]. It mainly refers to the efflux of drugs out of cancer cells through different membrane transporters, thus resulting in decreased intracellular accumulation of anticancer drugs and chemotherapy failure. Numerous membrane transports which control the transport of different substrates into and out of the cell have been identified. The two major superfamilies are (1) ABC transporters, which are further divided into P-Glycoprotein (P-gp), breast cancer resistance protein (BCRP) and multidrug resistance-associated proteins (MRPs) and (2) solute carrier (SLC) transporters, which include organic anion transporters, organic cation transporters and organic anion transporting polypeptides [113][115]. To date, 48 ABC transporters have been identified in the human genome, which are further subdivided into seven families (A–G). ABC transporters are responsible for facilitating the efflux of excessive intracellular drugs, thus giving rise to a significant impairment of chemotherapeutic effects [116][117]. The ABC transporter families play major roles in chemoresistance arising in CRC, due to their overexpression [113][114][115]. For instance, overexpression of the ABCB1 efflux transporter gave rise to 5-FU resistance in CRC cell lines [118], while most of the ABC transporters are responsible for DOX resistance [119]. Furthermore, upregulation of MRP2 and BCRP was also involved in the cisplatin-induced drug resistance in CRC cell lines [120], while overexpression of MRP2 was reported to be responsible for cisplatin resistance in patients undergoing chemotherapy and for OXA resistance in CRC cell lines [121].
Non-transport-based mechanisms can also give rise to resistance in CRC and are often linked to altered activities of specific enzymes and alteration in different signalling pathways and cell death pathways. Apoptosis and autophagy are two programmed cell death pathways triggered by the cells. Apoptosis is initiated via different extracellular and intracellular signals while autophagy is initiated due to different stressful stimuli [113]. Evasion of apoptosis, one of the hallmarks of human cancers, contributes to carcinogenesis and tumor progression, as well as drug resistance in cancer. Resistance to apoptosis in cancer cells, in this cases CRC, is linked to increased expression of antiapoptotic genes and proteins, as well as decreased expression of pro-apoptotic genes and proteins [122]. Thus, Bcl-XL, Mcl-1, B-cell Lymphoma (Bcl-2) and X-linked inhibitor of apoptosis tend to be overexpressed in CRC, while Bax, p53, Bim and apoptotic protease activating factor 1 are mutated or suppressed [122]. This was in fact proven by different research groups, for example CRC cell lines were resistant to DOX and apoptosis due to the Bcl-2 protein level being significantly upregulated as compared to the parental cell lines while the expression of p53 and BAX were significantly downregulated [123]. Moreover, loss of Bax expression was shown to decrease the sensitivity of HCT116 cells to apoptosis induced by 5-FU and OXA [113]. The role of autophagy in tumorigenesis is still controversial. Autophagy can promote the survival of rapidly growing CRC cells by targeting damaged or aged organelles for degradation and recycling [113]. At later stages of carcinogenesis, autophagy may act to stimulate tumor expansion by providing energy and nutrients important to the metabolism and growth of malignant cells, or by increasing drug resistance [115]. The p38MAPK pathway plays a key role in autophagy, especially in cellular responses to 5-FU. Studies reported that the inhibition of this pathway relates with a decrease in 5-FU-mediated apoptosis, stimulating CRC cell resistance. 5-FU resistance mediated by p38MAPK pathway inhibition is linked to autophagic response as it induces a decrease in p53-driven apoptosis without effect on p53-dependent autophagy. Consequently, the p38MAPK signaling pathway plays a critical role in CRC cell 5-FU resistance by controlling the balance between apoptosis and autophagy [124].
Apart from the cell death pathways, chemoresistance in CRC is also related to signaling pathways common to many other cell events. Thus far, several studies have indicated the relationship between different signaling pathways with chemoresistance of CRC cells. The Epithelial Growth Factor Receptor- Rat Sarcoma- Mitogen Activated Protein Kinase (EGFR-RAS-MAPK), Vascular Endothelial Growth Factor/Vascular Endothelial Growth Factor Receptor (VEGF/VEGFR), Phosphatidylinositol-4,5-bisphosphate 3-kinase/Protein Kinase B (PI3K/AKT), WNT/β-catenin, Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and Notch1 signaling pathways seem to be the most important signaling pathways involved in chemoresistance of CRC cells [125][114]. The most common genetic changes associated with CRC progression are mutations that deregulate the Wnt/β-catenin signaling cascade. This pathway is essential for maintaining cell homeostasis and embryonic development, and is associated with tumor cell proliferation, apoptosis, invasion, stemness and chemotherapy resistance [126]. The AKT/PI3K signaling pathway is activated uncontrollably in CRC due to mutations in various components of this pathway, as well as mutations in inhibitors such as PTEN, which enhance chemoresistance [106][114][127][128]. The function of PTEN is to regulate the activity of PIP3 by dephosphorylation, and thus inhibit Akt activity. PTEN mutation and loss of its activity thus leads to constitutive activation of Akt, promotion of cell survival and enhances chemoresistance [106]. The VEGF/VEFGR pathway is related to angiogenesis and lymphangiogensis in tumour growth. The VEGF family consists of five members (VEGF-A, -B, -C, and -D and placental growth factor (PIGF)), which can bind to endothelial cells via tyrosine kinase VEGF receptors. Vascular endothelial growth factor receptors (VEGFRs) are split into three, VEGFR-1, -2, and -3, along with the non-tyrosine kinase coreceptors neuropilin-1 (NP-1) and NP-2 [125]. The VEGF family members can interact with different proteins to regulate angiogenesis. Over-expression of the VEGF gene and high levels of circulating VEGF protein are both associated with worse prognosis in CRC [106][125]. NF-κB is a ubiquitous transcription factor that mediates a cytoplasmic/nuclear signaling pathway and regulates gene expression of various cytokines, cytokine receptors and adhesion molecules involved in inflammatory and immune reactions. In addition, there are links between the initiation of NF-κB and control of apoptosis, proliferation, differentiation, migration, angiogenesis, and resistance to chemo and radiotherapies in CRC [129]. The Notch signalling pathway is involved in the maintenance and homeostasis of intestinal epithelium and controls the cellular fate of intestinal stem cells and differentiation of colonic goblet cells. In CRC, this pathway is critical for maintaining the balance between cell proliferation, apoptosis, and differentiation, and thus its dysregulation results in further progression of CRC [130]. In fact, dysregulation of the Notch signaling pathway contributed to CRC progression, metastasis, and inhibition of apoptosis [131]. Lastly, the EGFR-RAS-MAPK pathway participates in many cellular processes, including the growth, proliferation, and survival of normal cells. If dysregulated, it modulates the growth, survival, proliferation, metastasis and chemoresistance of neoplastic colorectal cells [128].
Some of these signalling pathways have been shown to display a relationship between the expression of ABC transporters, particularly the P-gp transporter and certain pathways. For instance, there is a relationship between P-gp and the NF-κB signaling pathway, and it seems that inhibiting this signaling pathway leads to P-gp downregulation [132]. In addition, the AKT/PI3K signaling pathway and P-gp upregulation have been shown to contribute to 5-FU resistance in CRC cell lines [133]. The Wnt/β-catenin signaling cascade also contributed to enhanced resistance to various chemotherapeutic agents in CRC through upregulating MDR1 as reviewed by Yuan et al. [126]. Apart from interacting with ABC transporters, the signalling pathways can crosstalk between themselves. For example, a study has shown the relationship between the Notch signaling pathway and 5-FU and OXA resistance in CRC cell lines [134]. The Notch signaling pathway exerts its effect on chemoresistance by interacting with the WNT/B-catenin signaling pathway [134]. Downstream target genes in the Wnt/β-catenin signaling cascade have been shown to regulate drug resistance by controlling apoptosis. For instance, MMP-7 (matrilysin, a metalloproteinase with prometastatic function) could increase the OXA resistance of colon cancer cells by decreasing the Fas receptor that stimulates cell apoptosis [126][135]. Activation of VEGFR-1 under pathological conditions mediates the activation of several downstream pathways, such as PI3K/AKT/MAPK/ERK, while the VEGFR-2 interacts with VEGF-A [125]. Upon activation of VEGFR-2, tyrosine residues are phosphorylated together with the initiation of different pathways, such as RAS/RAF/ERK/MAPK pathways, by which epithelial cell growth is promoted, and the PI3K/AKT pathway, by which cell apoptosis may be avoided and chemoresistance is promoted [125].
EMT, a process that drives a cellular trans-differentiation continuum under physiological conditions and pathological states, has also been shown to be one of the main reasons why cancers become resistant to the treatment being administered. During EMT, epithelial cells progressively lose their typical morphological features (cell polarity, cell-to-cell contacts, membrane adhesion) and develop a mesenchymal phenotype, with the typical cellular stellate morphology, different propensity for intercellular signaling, as well as overall distinct cytological and tissue architecture [115]. EMT in CRC malignant colonocytes may be induced by different stimuli arising from external sources such as transforming growth factor β (TGF-β), plus various cytokines working together with intracellular operative signalling cascades including PI3K and NF-κB, along with other stimuli [115][136]. However, EMT can also be induced by different cytotoxic drugs which are typically used for CRC. For example, CRC cell lines treated with DOX induced EMT cell phenotypes, TGF-β signaling, and a significant increase in P-gp expression, which gave rise to resistance [119][137]. Similarly, 5-FU-resistant CRC cell lines showed an increase in different mesenchymal markers, together with an increase in EMT-inducing transcription factors Zeb2, Zeb1 and Twist [138].
As previously mentioned, most of the chemotherapeutic drugs being discussed in this review function by interfering with the synthesis of RNA and DNA. Thus, DNA damage repair mechanisms can become an important contributor to drug resistance. The mismatch repair system (MMR) is a replication fidelity complex which is responsible for detecting and repairing any faults or mismatched bases arising in the cell throughout DNA replication and various other processes [139]. In CRC, mismatch repair deficiency (dMMR) can occur in both sporadic and hereditary CRC [112]. Germline mutations in either MutL homolog 1 (MLH1), MutS homolog 2 (MLH2), PMS1 Homolog 2 (PMS2), or mutS homolog 6 (MSH6) give rise to dMMR [112][140]. Epigenetic hypermethylation of the MLH1 promoter can also give rise to dMMR [139][141]. Different studies have been carried out to understand the relationship between MMR and the response of different CRC therapeutic agents in the development of resistance. It has been shown that cells which have dMMR tend to be resistant to certain cytotoxic agents which function by targeting the DNA. For instance, DNA mismatches produced by incorporation of 5FdUTP into DNA were not detected in dMMR cells, which result in cell survival and 5-FU resistance, while chemoresistance was reversed once MMR deficiency was corrected [140][142]. This phenomenon was also observed when using cisplatin in the study carried out by Martin et al. [143], while loss of either MSH2 or MLH1 function resulted in resistance to DOX [144]. Despite OXA being a platinum compound like cisplatin, OXA resistance in dMMR CRC cells lacking MMR is highly unlikely, as studies have shown that cisplatin–DNA adducts can be recognized and repaired by the mismatch repair system (MMR), whereas OXA–DNA adducts are not [145][146]. Thus, cells which have dMMR are intrinsically resistant to cisplatin but remain sensitive to OXA. However, OXA resistance in CRC can arise due the excision repair cross-complementing group 1 (ERCC1), and its catalytic partner Xeroderma Pygmentosum group F (XPF), which are key elements for the nucleotide excision repair (NER), responsible for repairing DNA adducts arising due to platinum compounds [147][148].
Lastly, based on the drugs being administered, chemoresistance in CRC can also arise due to drug metabolism and specific targets/enzymes [112][149][106]. These two modes of resistance tend to be specific to the drug being administered, since different chemotherapeutics are targeted or metabolised by different targets/enzymes. For example, 5-FU resistance tends to also arise due to malignant cells overexpressing dihydropyrimidine dehydrogenase (DPD), which is the enzyme responsible for the first step of 5-FU catabolism [149][106]. In fact, high levels of DPD mRNA expression in CRC cells have been associated with 5-FU intrinsic resistance [106][150][151]. On the other hand, if a low expression of DPD is present, 5-FU cannot be metabolised efficiently [112].
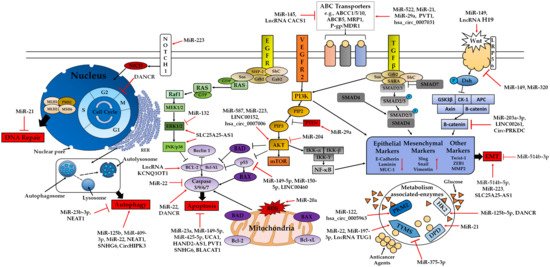
Figure 2. Classic mechanisms involved in drug resistance (5-FU, OXA, Cisplatin and DOX) in CRC. miRNAs, lncRNAs and circRNAs modulate drug resistance in CRC through different pathways such as the PI3K/AKT, EGFR-RAS-MAPK, WNT/β-catenin, NF-κB, TGF-β and Notch1 signaling pathways, cell death pathways (apoptosis, autophagy), EMT, DNA repair and ABC transporters. Arrows represent activation effect, and the ‘T’ symbols represent inhibition. Arrows and ‘T’ symbols in red represent some of the different ncRNA targets involved in chemoresistance to be discussed in this review. Abbreviations: MLH1: MutL homolog 1; MLH2: MutS homolog 2; PMS2: PMS1 Homolog 2; MSH6: MutS homolog 6; NICD: Notch intracellular domain; RER: Rough Endoplasmic Reticulum; GRB2: Growth Factor Receptor-bound protein 2; SOS: Son of Sevenless; SHP-2: SH2 domain-containing protein tyrosine phosphatase-2; GAB2: GRB2 Associated Binding Protein 2; SHC: SH2 containing protein; GSK3β: Glycogen Synthase Kinase 3β; AKT: Protein Kinase B; mTOR: Mammalian target of rapamycin; Iκκ: IκB kinase; Dsh: Dishevelled; PIP2: Phosphatidylinositol 4,5-bisphosphate; PIP3: Phosphatidylinositol (3,4,5)-trisphosphate; APC: Adenomatous Polyposis Coli; HK2: Hexokinase 2; TYMS: thymidylate synthases; DPD: Dihydropyrimidine Dehydrogenas; PKM2: Pyruvate Kinase 2; SARA: SMAD anchor for Receptor Activation.