1000/1000
Hot
Most Recent

Microbial symbionts are nowadays considered of pivotal importance for animal life. Among the many processes where microorganisms are involved, an emerging research avenue focuses on their major role in driving the evolution of chemical communication in their hosts. Volatiles of bacterial origin may underlie chemical communication and the transfer of social information through signals, as well as inadvertent social information.
Interactions between animals and their associated microorganisms (i.e., microbiota) are nowadays considered of pivotal importance to understand the physiology, morphology, and behavior of animals, as well as the outcomes of their interactions with abiotic and biotic environmental conditions [1]. Beyond pathogenesis, the most commonly studied effects of microorganisms on animals are those that link the gastrointestinal microbiota with facilitation of nutrient absorption, or even the synthesis of some essential micronutrients [2][3][4]. During the last two decades, the scientific interest has begun to consider the microbiota as an essential component of living animals, therefore affecting their evolution [5]. An emerging topic in evolutionary biology deals with the importance of the microbiome in mediating communication in their host organisms [6].
Animals acquire information from the environment by direct interactions in trial-and-error-tactics (personal information), or by monitoring the interactions of others with the environment and their outcomes, thereby acquiring what is called social information (SI) [7]. Social information can be based on signals, which are traits that specifically evolved to convey information to receivers [8]. Alternatively, social information can also be based on cues provided inadvertently by individuals while engaged in their biological activities (inadvertent social information, ISI) [7]. Signals usually inform or advertise receivers on the phenotypic condition and capabilities of the sender, which supposedly benefits both sender and receiver [8][9]. ISI may inform bystanders, for instance, about resource location, but also about the quality of the resource, which is revealed by the performance or phenotypic quality of the cue sender (i.e., public information) [7]. Importantly, signals, as well as ISI, are supposed to reliably convey information on the phenotypic condition of the sender. Honesty of signaling characters has mainly relied on the hypothesis that only high-quality individuals will be able to afford its associated costs [8][10][11]. Instead, ISI is supposed to convey information on the phenotypic quality of the sender as a result of individual performance [7].
Depending on the type of the sensitive channel used to transmit the signal or to gather ISI, stimuli have been mainly classified as visual, auditory, or chemical. The use of chemicals is the most ancient, widespread, and shared way used by living organisms to evaluate their environment and to communicate with each other [12]. Remarkably, symbiotic bacteria, or what as a whole is known as microbiota, are largely responsible for animal scents [13][14]. The role of symbiotic bacteria in animal chemical communication is therefore paramount [6]. Moreover, the microbiota is intimately related to the phenotypic quality and physiological activity of their animal hosts [15][16][17] by influencing their growth and development [1]. This indeed will affect characteristics of signals and ISI that conspecifics and heterospecifics could use. Particularly interesting is the possibility that chemical signals and cues of bacterial origin can be eavesdropped on by unintended receivers, such as parasites and predators, when locating and selecting hosts and prey.
Yet symbiotic microorganisms may also influence the outcomes of the interactions between their hosts and their hosts’ enemies (predators and parasites) in other ways. For instance, microorganisms largely determine host health and condition [1][3], and these effects could be also used by predators and parasites as inadvertent social information that facilitate host detection and/or selection [14]. Symbiotic microorganisms can also produce metabolites with antimicrobial properties [18] that clear or prevent parasitic infections. Some bacterial symbionts are also known to produce metabolites that deter predators or parasites [19]. Symbiotic microorganisms might even be related to adaptive hormonal and immunological plastic responses of hosts against stressful environmental conditions, including those related to the risk of parasitism or predation [20]. All these possibilities highlight the hypothetical role of microorganisms in driving the interaction between hosts and their parasites and predators, and we have here reviewed current knowledge on these matters.
The role of microbial symbionts on animal chemical communication has been reviewed several times during the last decade [6][14][21] and was not the aim of this essay. Here, we rather sought to explain the rationale behind the social information value of volatiles of microbial origin in scenarios of animal communication. We also aimed to formulate key predictions to assess this hypothetical role of bacterial symbionts, and discuss the importance of bacterial symbionts in the evolution of conspecific communication, and in host–parasite and prey–predator interactions. The animal microbiome may cause beneficial as well as detrimental effects to its host. Special attention has been paid to the possibility that host enemies might eavesdrop on inadvertent social information mediated by beneficial microbiotas from their victims. An overview of the potential interactions between hosts and their bacterial symbionts in scenarios of social communication, parasitism and predation that are dealt with in this essay is shown in Figure 1.
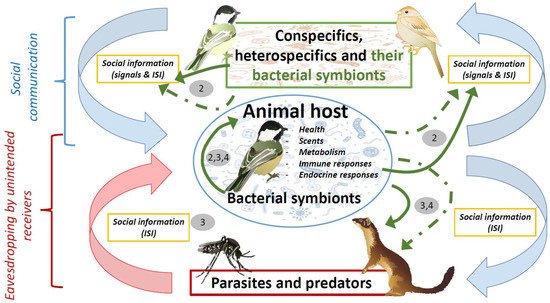
Figure 1. Diagram showing hypothetical influence of bacterial symbionts (green arrows) in scenarios of social communication, parasitism, and predation. These influences could be directly due to either bacterial metabolism or products with antimicrobial or antipredatory properties (solid arrows), or indirectly through their effects on host characteristics (i.e., health, scents, metabolism, immunity, and hormones) (dashed green arrows). Bacterial symbionts contribute to social information that is received by conspecifics or heterospecifics, including parasites and predators. The negative effects of parasites and predators (red arrow) would be directly counteracted by defensive products of bacterial origin, or indirectly by host defensive traits that are also influenced by bacteria (continuous and dashed green arrows connecting the host with parasites and predators). These negative effects however will be enhanced by eavesdropping on inadvertent social information directly or indirectly mediated by host microbial symbionts and, thus, parasites and predators will also influence the symbiotic association between animals and microorganisms. Pathogenic parasites could also influence health and, consequently, bacterial symbionts of their victims, and, thus, parasites could indirectly affect conspecific communication. Numbers refer to main sections in the text where that relationships are covered. Symbols courtesy of the Integration and Application Network, University of Maryland (ian.umces.edu/symbols/) and freepik.com (accessed on 5 February 2021).
The hypothetical role of microorganisms in animal communication is rooted in the “fermentation hypothesis”. This hypothesis was originally formulated to explain the odors of anal sac secretions of cats and foxes in the 1970s [22][23], but is now applied to the general odor profile of animals that could operate in a large variety of scenarios of olfactory communication [6][21][24][25]. Until very recently, microbial production of chemical signals had been mainly described in mammals and insects [6]. However, solid evidence for the role of bacterial symbionts in producing volatile metabolites that contribute to the host odor profile is rapidly being accumulated for a wider range of animal taxa, including not only mammals [15][16] and insects [14][26][27], but also amphibians [28] and especially birds [17][29][30].
Evidence supporting the key role of bacterial symbionts in animal communication represents one of the most fascinating and important advances that chemical ecology has experienced during the past few years [21]. The role of bacterial symbionts in animal communication is based on the assumption that some genetically and environmentally determined characteristics of animals, like those related to diet and immunity, also determine their microbial symbionts [31][32]. Consequently, volatiles of microbial origin would inform the characteristics of their animal hosts (social information), and therefore, these chemicals would contribute to olfactory communication [6]. For instance, because of the many factors determining animal microbiotas, and the enormous variability of associated microbial volatiles, the particularities of chemical profiles due to the metabolism of bacterial symbionts may communicate individual identity to conspecifics (i.e., an individual signature) [21][33]. Microbial volatiles may also aid in easy navigation towards nest locations for parents or offspring due, for instance, to the particular volatile profile of feces around nests [34][35]. Also, in scenarios of parent–offspring communication, volatiles from symbiotic bacteria might help newborn mammals to recognize their own mother’s nipples [36][37], while in sexual selection scenarios these volatiles can be used to choose a genetically compatible partner [38][39].
Previous research on this topic has been mainly focused on the possibility that volatiles derived from the metabolisms of host bacterial symbionts function to assess host quality by conspecifics. As we mentioned before, the microbiota composition, and thus volatile profiles of bacterial origin, depends on host characteristics, which include components of the host phenotypic quality. Thus, because these volatiles would convey valuable information to conspecifics, selection will favor receivers using such cues of microbial origin [31]. However, to demonstrate that microorganisms convey information on the phenotypic characteristics of their hosts, linking particular volatiles predicting host characteristics with the microorganisms that produce such volatiles is needed. Exploring associations between microbiotas and the odor profiles of the hosts reflecting their physiological characteristics is nowadays a fruitful area of research that will help to unveil general patterns on the role of microbiotas in social communication. Metagenomics, together with other omic techniques studying the metabolic production of microbial communities (e.g., proteomics and metabolomics), will help to characterize the microorganisms responsible for the production of particular volatile metabolites [40], allowing us to fill important gaps in our knowledge regarding the role of the animal microbiome in chemical ecology and communication.
The evolution of chemical signals mediated by microbial symbionts requires, not only that those odors reliably reflect characteristics of their hosts, but also that receivers use the conveyed information, and that such communication benefits both sender and receiver. By providing microorganisms with substrates in special locations such as the gut or glands, animals of several taxa cultivate bacteria, producing substances that are valuable for them in terms of micronutrient provisioning, antimicrobial defenses, or even signaling [1][41][42]. Scents originating from symbiotic microorganisms that inhabit animal glands are particularly important to gain insight into the evolution of microbially mediated chemical communication. This is mainly because most scents derived from animal glands have been traditionally considered as classical examples of chemical signals. Evidence supporting the existence of microbial symbionts growing within such scent glands are being accumulated in the literature, especially in exocrine glands, such as the anal glands of mammals [15][16] and the uropygial glands of birds [17][30]. Similarly, scents derived from microorganisms that enhance the survival and reproductive success of their hosts, such as those from gut microbiotas, would also signal host phenotypic quality, therefore evolving into an associated signaling role. Consequently, in these cases, where scents of microbial origin are part of the animal chemical signaling, the evolution of chemical communication should entail changes not only in genetically inherited characteristics of hosts, but also in characteristics of their microbial symbiotic communities. However, changes in animal hosts and in their microbiotas should not be seen as completely independent, because animal characteristics will largely determine characteristics of their microbiotas.
Even though it is generally assumed that the fitness of the host is often linked to that of its microbiota [43][44], but see [45], the evolution of symbiotic bacterial communities by means of natural selection acting on hosts entails important theoretical challenges. This is mainly because the characteristics of microbial symbionts are not directly determined by animal genomes and, thus, natural selection acting on host performance or fitness would not be able to directly modulate the bacterial community of symbionts, nor their chemical profiles. To overcome this theoretical problem for explaining the evolution of host microbiomes, some authors have claimed that microbiomes and individual hosts should be considered together as the unit (holobiont) where natural selection acts [46][47][48]. It has been broadly recognized that the characteristics and composition of host-associated microbial communities parallel the phylogeny of the related host species, and the holobiont concept would a priori help to understand the evolution of such phylo-symbiosis [49]. This approach, however, may entail some other theoretical problems related to group selection theory [50]. A main critique to the holobiont concept is that fitness of hosts and symbionts are not fully linked, especially not for all members of a host-associated microbiota [45]. Thus, alternative scenarios explaining phylo-symbiosis have been proposed and explored. One possibility is to consider genetically determined traits in animal hosts that allow or favor maintenance of certain microbial communities, driven, for instance, by host variability in diet or habitat. The allelic variation in genes determining such traits will therefore predict the composition of, or functional variation in, microbiotas [51]. In this case, host characteristics determining, for instance, the mode of transmission of bacterial symbionts and the characteristics of the environment where microbes are hosted will also govern the composition of symbiotic bacterial communities and their metabolic activity [52]. Therefore, it is possible that natural selection acting on hosts could determine the metabolic activity of the microbiota, including those microorganisms responsible for the production of volatiles with importance in chemical communication. Similarly, for visual traits, we know, for instance, that the eggshell coloration of hoopoes (Upupa epops) is affected by uropygial secretion rubbed on eggshells by females during incubation, which is indeed mediated by symbiotic bacteria hosted in the uropygial gland [53]. The eggshell color functions as a signal of female quality [54], while the symbiotic bacterial community hosted in the uropygial gland of hoopoes have a significant genetic component [55][56]. Thus, because characteristics of the microbial community of the uropygial secretion are likely mediated by physiological characteristics of hosts (where natural selection can operate), natural selection processes would also be responsible for egg coloration in hoopoes.
Similar processes to those described above for hoopoes can also operate for olfactory traits mediated by symbiotic microorganisms. Host characteristics that enhance the establishment of the microbiota with direct beneficial effects for hosts could be identified by characteristic host odors. Moreover, because of the potentially narrow link between those host traits and characteristics of the volatile profile of the associate microbiota, host odor mediated by bacterial symbionts will also reflect host characteristics favoring the establishment of particular microbiotas. Thus, the effects of natural selection acting of host traits could easily be tracked by following variation in host odor profiles. Interestingly, because mating with individuals with characteristics that enhance growth of beneficial microorganisms would be of selective advantage, sexual selection acting on olfactory traits mediated by bacterial symbionts will also accelerate the evolution of these characteristics. Future research should focus on identifying (i) physiological or morphological host traits enhancing the establishment and growth of beneficial microbiotas, (ii) characteristic microbial volatiles narrowly reflecting potential fitness effects for their hosts, and (iii) whether sexual selection favors hosts of particular bacterially mediated odor profiles. These research will allow to gain insight into the mechanisms underlying the evolution of hosts characteristics that favor particular microbiotas.
Microbial symbionts would also contribute to the inadvertent social information provided by their hosts. Interestingly, this information does not necessarily benefit hosts, but reliably informs conspecifics and heterospecifics about host phenotypic characteristics or condition. It may be the case that pathogens, or parasite infections, influence the host microbiota, which would result in animals displaying particular volatile profiles. Conspecifics could thus use that ISI as a warning chemo-sensory signal to, for instance, avoid close contacts with sick individuals. For example, in humans, experimental activation of the immune system affected body odor, which was judged by conspecifics as less pleasant, more intense, and less healthy [57]. Similarly, mice, mandrills, and lobsters are able to identify sick conspecifics via chemical cues [58][59][60][61]. Thus, the role of bacterial symbionts in indirectly mediating communication of health status to conspecifics could be widespread among animals. Importantly, emitting such volatiles might have negative effects on their hosts. Therefore, there may be evolutionary processes to selecting host traits that shape the microbiota in emitting volatiles with less detrimental effects. Future work should also explore the role of bacterial symbionts as producers of inadvertent social information with detrimental effects for their hosts.