1000/1000
Hot
Most Recent

G-protein coupled receptors (GPCRs) are membrane proteins that convey extracellular signals to the cellular milieu. They represent a target for more than 30% of currently marketed drugs.
G-protein coupled receptors (GPCRs) are membrane proteins that pass on extracellular signals to the cell using heterotrimeric GTP-binding proteins (G-proteins). GPCRs are integral membrane proteins that possess seven transmembrane α-helices (denoted TM1 to TM7) connected with three intracellular (IL1 to IL3) and three extracellular (EL1 to EL3) loops (Figure 1A). The cysteine in the middle of ECL2 forms disulfide bridge with cysteine at the edge of TM3. The N-terminus of GPCR is oriented out of the cell and may be glycosylated at asparagine or glutamine residues. The C-terminus is oriented to the cytoplasm and may be palmitoylated or myristoylated at cysteine residues. Individual amphiphilic TMs form a circular bundle with a hydrophilic pocket among them that is accessible from the extracellular side (Figure 1B). The pocket serves as an orthosteric site for endogenous transmitter or hormone. More than 30% of currently marketed drugs act at GPCRs and thus GPCRs represent a very important pharmacological target [1].
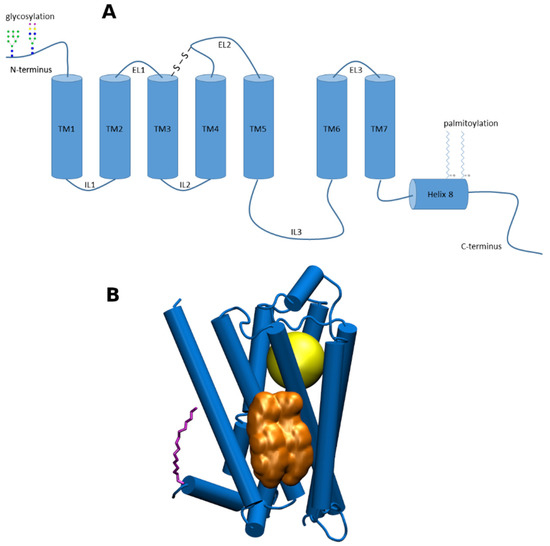
Figure 1. (A), a schematic representation of structural features of G-protein coupled receptors (GPCRs) of class A; (B) 3D representation of GPCR of class A. Yellow—the orthosteric binding site; gold—cholesterol dimer bound to cholesterol consensus motif (CCM); purple—palmitic acid covalently bound to cysteine in Helix 8.
Most of neurotransmitters act at several receptor subtypes of a given receptor family. This divergence allows one signalling molecule to elicit different cellular responses depending on the distribution of the receptor subtypes in the body. For a pharmacological agent to target body organs selectively, it has to be able to differentially influence the activation of individual receptor subtypes. In general, the binding site for a given endogenous signalling molecule is conserved among its receptor subtypes. This is necessary for accommodating the signalling molecule during the evolution of receptor subtypes. The sameness of the orthosteric site, however, makes finding subtype-selective compounds acting at the orthosteric binding site extremely difficult. In contrast to orthosteric sites, secondary allosteric binding sites on receptors are not under such evolutionary pressure and vary among subtypes [2]. Therefore, a lot of effort was given to the research of allosteric binding sites and allosteric modulators. A large number of various allosteric modulators of GPCRs that bind to the extracellular or intracellular domains were identified.
Cholesterol (CLR) is a sterol-like type of lipid. CLR composes about 30% of all animal cell membranes. The primary function of CLR is structural. It regulates membrane fluidity. Other non-structural functions of CLR include its physical interaction with many membrane proteins including GPCRs (Figure 1B). This interaction results in alteration of receptor properties in terms of the processes of ligand binding, receptor activation and signal transduction [3][4]. Thus, membrane CLR can be considered an allosteric modulator of GPCRs possessing its own specific allosteric binding site.
CLR is a polycyclic and amphiphilic molecule that is found in high abundance in cell membranes. Its main function is regulation of membrane fluidity by facilitation of the formation of ordered phases in the lipid bilayer via composite interactions between lipid components. CLR is a shorter and more rigid molecule in comparison with phospholipids. Therefore, parts of the membrane close to CLR molecules are more rigid and thinner. Fluidity and thickness of the membrane, in turn, affect membrane protein trafficking.
CLR has a flat asymmetric structure defined by a planar α-face and rough β-face, named according to the nomenclature of ring compounds [5]. CLR in the membrane may exist as monomer or form dimers oriented α-face to α-face, the so-called face-to-face dimers, stabilized by Van der Waals contacts (Figure 2A) [6]. Face-to-face CLR dimers were found in X-ray crystal structures of membrane proteins. Another type of CLR dimer may be stabilized by a hydrogen bond between hydroxyl groups (Figure 2B). However, CLR hydroxyl group rather interacts via hydrogen bonding with other membrane lipids or proteins [7]. The third type of CLR dimers, trans-bilayer tail-to-tail dimer, has been hypothesized to exist in membranes (Figure 2C) [8].
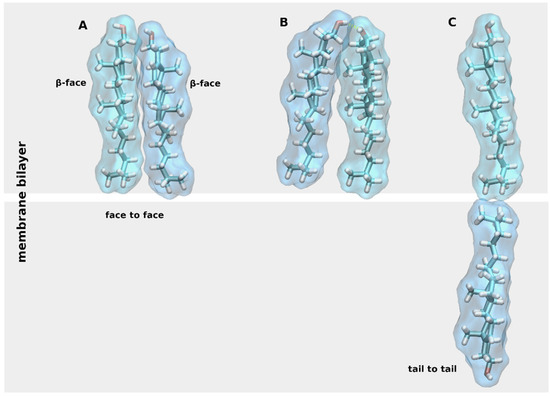
Figure 2. Cholesterol (CLR) dimers. Three types of CLR dimers: (A), face-to-face dimer; (B), dimer stabilized by a hydrogen bond (yellow dashed line); (C) tail-to-tail dimer.
In principle, CLR may affect GPCRs in two ways. It may either directly bind to the receptor and thus allosterically modulate the affinity of ligands, efficacy of agonists and spontaneous activity of the receptor. Alternatively, CLR may affect GPCRs indirectly by changing fluidity and organization of the membrane that in turn affects signalling of GPCRs. Direct modulation of GPCRs by CLR requires its interaction with specific sites on receptors with sufficient affinity. Such sites were identified in many GPCRs; see below. In contrast, the indirect mechanism does not involve CLR-specific binding site. As stated above, CLR decreases membrane fluidity that slows down the diffusion of solute molecules like receptors, channels or membrane enzymes that in turn slows-down kinetics and decreases the efficacy of signal transmission from a given receptor to its effector. In membranes rich in CLR content, CLR has a propensity to associate into patches. High CLR content increases the order of neighbouring acyl chains that leads to increased bilayer thickness [9]. These membrane microdomains are termed lipid rafts and substantially affect signal transduction [10]. The hydrophobic mismatch is defined as the difference between the hydrophobic membrane thickness and the peripheral length of the hydrophobic part of the membrane-spanning protein [11]. The membrane-perpendicular length of GPCRs is shorter in an inactive conformation than in an active conformation. Therefore, GPCRs in an inactive conformation may be preferentially sorted to non-raft regions that represent a thinner part of the membrane. Consequently, keeping a receptor in the non-raft region may constrain it in an inactive conformation [12]. Thus, keeping a receptor in non-raft region ablates its signalling. Moreover, the differential localization of proteins in various microdomains increases the specificity of signalling. Co-localization of several signalling pathways at a given microdomain, for example, may promote the formation of a signalling hub that enables integration of distinct signalling pathways at the receptor-membrane interface [13][14]. Thus, lipid rafts play a unique role in cell physiology and pathology and represent possible target in hematopoietic, inflammatory, neurodegenerative, and infectious diseases [15]. Taken together, the indirect effects of membrane CLR are diverse and bring complexity to GPCR signalling.
CLR was found co-crystallized with many GPCRs of class A suggesting possible specific binding. At the time of writing of this review, 44 X-ray or cryo-EM structures of 18 receptors of GPCRs of Class A have been published in the RCSB database (https://www.rcsb.org/, accessed on 20 January 2021) (Table 1). CLR was found co-crystallized with receptors for structurally different agonists including biogenic amines like adrenaline (α2C, β2) or serotonin (5-HT2B), peptides like angiotensin (AT1), chemokines (CCR9, CXCR2, CXCR3), endomorphins (κOR, μOR), endothelin (ETB), formyl peptide (FPR2) or oxytocin (OTR), purines like adenosine (A2A) or ADP (P2Y1, P2Y12), endocannabinoids (CB1, CB2), and eicosanoids like leukotriene (CLT2). CLR in crystals appeared as monomer or dimer. For some receptors, CLR binding was confirmed in several crystal structures, e.g., β2, 5-HT2B, or A2A. On the other hand, for some receptors, X-ray structures provide contradictory results, e.g., AT1, CXCR2 or ETB. It also should be noted that in some cases CLR was found in an unexpected orientation, for example, parallel to membrane (CB2) or hydroxy group in the middle of membrane bilayer (CB1). Additionally, no CLR was found in the crystal structures of GPCRs at which CLR was shown to have a profound effect on ligand binding or receptor activation; see below. Thus, information on the interaction of CLR with GPCRs inferred from crystal structures should be taken with caution. Further, cholesteryl hemisuccinate (CHS) that is used for solubilisation of biological membranes was found co-crystallized with GPCRs. CHS may compete out CLR from binding to GPCR. As it is not certain whether co-crystallized CHS indeed binds to the CLR-specific binding site on GPCRs or is the result of the solubilization process, CHS binding to GPCRs is not covered in this review.
Table 1. Cholesterol in crystal and cryo-EM structures. List of X-ray and cryo-EM (blue PDB codes) structures of Class A GPCRs containing cholesterol. Rec.—receptor subtype, Code—PDB ID code, G-prot.—a subclass of G-proteins mediating the primary response, Conf.—active or inactive conformation of the receptor, CLR—monomeric or dimeric state of CLR, Leaflet—location of CLR in the inner or outer leaflet of the membrane, TM—transmembrane helices CLR is interacting with, Ref.—reference.
|
Rec. |
Code |
G-prot. |
Conf. |
CLR |
Leaflet |
TM |
Notes |
Ref. |
|---|---|---|---|---|---|---|---|---|
|
α2C |
6KUW |
Gi |
Inactive |
Monomer |
Out |
1, 7 |
cholesterol is a part of the protomer-protomer interface |
TBP |
|
β2 |
2RH1 |
Gs |
Inactive |
Dimer |
In |
2, 3, 4 |
[16] |
|
|
Monomer |
In |
1, 8 |
||||||
|
β2 |
3D4S |
Gs |
Inactive |
Dimer |
In |
2, 3, 4 |
[17] |
|
|
β2 |
6PS0 |
Gs |
Inactive |
Monomer |
In |
2, 3, 4 |
6PS2, 6PS3, 6PS4, 6PS5, 6PS7 same |
[18] |
|
κOR |
6PT2 |
Gi |
Active |
Monomer |
Out |
6 |
only with one protomer |
[19] |
|
κOR |
6B73 |
Gi |
Active |
Monomer |
Out |
6 |
[20] |
|
|
κOR |
6VI4 |
Gi |
Inactive |
Monomer |
In |
4, 5 |
only with one protomer |
[20] |
|
μOR |
4DKL |
Gi |
Inactive |
Monomer |
Out |
6 |
[21] |
|
|
μOR |
5C1M |
Gi |
Active |
Monomer |
Out |
6 |
[22] |
|
|
5-HT2B |
4IB4 |
Gq |
Active |
Monomer |
In |
1, 8 |
[23] |
|
|
5-HT2B |
5TVN |
Gq |
Active |
Monomer |
In |
1, 8 |
[24] |
|
|
5-HT2B |
6DRX |
Gq |
Active |
Monomer |
In |
1, 8 |
6DRY, 6DRZ, 6DS0 same |
[25] |
|
A2A |
4EIY |
Gs |
Inactive |
Dimer |
Out |
6 |
[26] |
|
|
Monomer |
Out |
2, 3, 4 |
||||||
|
A2A |
5IU4 |
Gs |
Inactive |
Dimer |
Out |
6 |
5UI7, 5UI8, 5UIA and 5UIB same |
[27] |
|
Dimer |
Out |
2, 3, 4 |
5UIB only monomer. |
|||||
|
AT1 |
6OS1 |
Gq |
Active |
Monomer |
In |
1, 8 |
6OS0 and 6OD1 no cholesterol |
[28] |
|
CB1 |
6N4B |
Gi |
Active |
Monomer |
In |
3, 4 |
[29] |
|
|
Monomer |
In |
3, 4 |
unexpected orientation with OH in the middle of the membrane |
|||||
|
CB2 |
6PT0 |
Gi |
Active |
Dimer |
Out |
6 |
[30] |
|
|
Monomer |
In |
5, 6 |
parallel to membrane |
|||||
|
Monomer |
In |
3, 4 |
parallel to membrane |
|||||
|
CCR9 |
5LWE |
Inactive |
Monomer |
In |
6 |
[31] |
||
|
CLT2 |
6RZ7 |
Gq |
Inactive |
Monomer |
In |
6 |
6RZ6, 6RZ9 dtto. |
[32] |
|
CXCR2 |
6LFM |
Gi |
Active |
Monomer |
In |
2, 3, 4 |
6LFO ditto, 6LFL no cholesterol |
[33] |
|
CXCR3 |
5WB2 |
Gi |
Active |
Dimer |
Out |
6 |
[34] |
|
|
ETB |
5X93 |
Gq |
Inactive |
Monomer |
Out |
1, 7 |
5XPR no cholesterol |
[35] |
|
FPR2 |
6LW5 |
Gi |
Active |
Monomer |
In |
6 |
[36] |
|
|
Monomer |
In |
2, 3, 4 |
||||||
|
FPR2 |
6OMM |
Gi |
Active |
Dimer |
Out |
1, 2 |
[37] |
|
|
Monomer |
Out |
6 |
||||||
|
Monomer |
In |
6 |
||||||
|
Monomer |
In |
3, 4, 5 |
||||||
|
OTR |
6TPK |
Gq |
Inactive |
Monomer |
Out |
4, 5 |
[38] |
|
|
P2Y12 |
4NTJ |
Gi |
Inactive |
Monomer |
Out |
1, 7 |
[39] |
|
|
Monomer |
In |
3, 4 |
unexpected orientation, binding to Y in DRY motif |
|||||
|
P2Y12 |
4PXZ |
Gi |
Inactive |
Monomer |
In |
2, 3, 4 |
[39] |
|
|
P2Y1 |
4XNV |
Gq |
Inactive |
Monomer |
Out |
2, 3, 4 |
[40] |
Several CLR sites can be distinguished (Table 1, Figure 3). A CLR dimer in the outer leaflet of membrane binds to A2A-adenosine receptor at a groove between TM2, TM3, and TM4 (Figure 3A) or TM6 (Figure 3C). A similar binding of CLR as in Figure 2A can be found at the P2Y1 receptor (4XNV) (Table 1). The κ-opioid receptor in an active conformation (6PT2, 6B73), the μ-opioid receptor at an active (5C1M) or inactive conformation (4DKL) and the CXCR3 receptor at an active conformation (5WB2) have CLR bound to the same site as the A2A-adenosine receptor shown in Figure 3C. A CLR monomer in the outer leaflet binds to the oxytocin receptor at TM4 and TM5 (Figure 3B) or to α2C-adrenergic receptor at TM1 and TM7 (Figure 3D). A similar binding site as in Figure 3D has been identified at the endothelin receptor (5X93) and purinergic P2Y12 receptor (4NTK) (Table 1). A CLR dimer in the inner leaflet of the membrane binds to the β2-adrenergic receptor in a groove formed by TM2, TM3, and TM4 (Figure 3E). The same CLR-binding site is present at the CXC receptor 2 (6LFM), formyl peptide receptor 2 (6CW5, 6OMM) and P2Y12 purinergic receptor (4NTJ). A CLR monomer in the inner leaflet binds to TM6 of an inactive conformation of the κ-opioid receptor (Figure 3F) or the CCR9 chemokine receptor (Figure 3G). A similar binding of CLR as in Figure 3G has been found at the cysteinyl leukotriene receptor 2 (6RZ7) and formyl peptide receptor 2 (6CW5, 6OMM). A special kind of CLR interaction at TM1 stabilized by palmitic acid covalently bound to cysteine in helix 8 can be found at the 5-HT2B receptor (Figure 3H) and also at the β2-adrenergic receptor (2RH1) and angiotensin receptor 1 (6OS1).
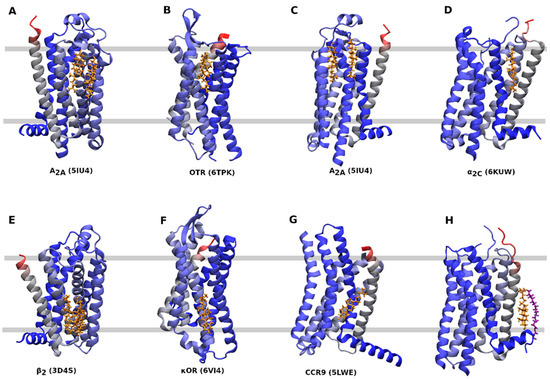
Figure 3. CLR binding sites. Orientation—extracellular side up, N-terminus—red, C-terminus— blue, cholesterol—gold, palmitic acid—purple. (A), CLR dimer binding to A2A-adenosine receptor (5IU4) at TM2, TM3 and TM4. (B), CLR monomer binding to oxytocin receptor (6TPK) at TM4 and TM5. (C) CLR dimer binding to the A2A-adenosine receptor (5IU4) at TM6. (D), CLR monomer binding to α2C adrenergic receptor (6KUW) at TM1 and TM7. (E) CLR dimer binding to the β2 adrenergic receptor (3D4S) at TM2, TM3, and TM4. (F), CLR monomer binding to the κ-opioid receptor (6VI4) at TM4 and TM5. (G), CLR monomer binding to the CCR9 chemokine receptor (5LWE) at TM6. (H), CLR monomer binding to the 5-HT2B receptor (4IB4) at TM1 and helix 8.
Receptors found co-crystallized with CLR mediate their primary functional responses via all three major subclasses of G-proteins: Gi, Gs, and Gq. None of the CLR-binding sites can be considered typical for a given receptor coupling pathway, suggesting that CLR-binding sites evolved independently from receptor coupling. Similarly, comparison of GPCRs in active and inactive conformations does not show any correlation with CLR binding. This suggests the absence of a common mechanism of CLR action on receptor activation.
Based on X-ray structures two putative cholesterol-binding motifs were postulated. Besides the so-called ’CLR recognition amino acid consensus’ (CRAC) domain common for all membrane proteins [41], the so-called ‘CLR Consensus Motif’ (CCM) was identified in the structure of the β2-adrenergic receptor (3D4S) [17]. CCM is the groove formed by 2 or 3 TMs. For the 3D4S structure, residues R151, L155 W158 in the TM4 and Y70 in the TM2 were identified as key CLR-binding residues (Figure 4). Although the orientation of the CLR dimer in the 2RH1 structure is slightly different from the 3D4S structure, key interactions with CCM are preserved [16]. The same applies to binding of the monomeric CLR in the 6PS0 structure [18]. In contrast, no CLR was found in four structures of the β2-receptor: 2R4R, 2R4S, 3KJ6, 3P0G. Based on bioinformatics studies of GPCR homology, the consensus sequence of CCM has been established as R/K-X5-I/V/L-X5-Y/W in the one helix and F/Y in the opposing helix. Residues R/K and F/Y of CCM are at the intracellular edge of TM helices. Residues Y/W are approximately in the middle of the membrane. The hydroxyl group of CLR interacts with a basic residue of CCM that are abundant at the intracellular edge of TMs. The β-face of the CLR dimer binds strongly with W or Y via hydrophobic, mainly π-π stacking, interactions. The CRAC domain (R/K-X5-Y-X5-L/V) and its reversed CARC (L/V-X5-Y-X5-R/K) are similar to the CCM in having R or K at the edge of the membrane. In comparison to CCM, positions of aromatic and hydrophobic residues are swapped in CRAC and CARC. While CRAC and CARC accommodate monomeric CLR, the CCM may bind a CLR dimer.
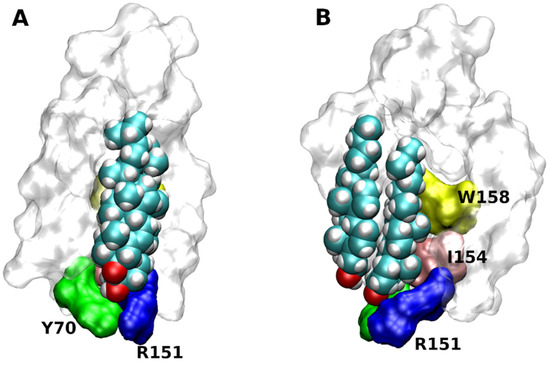
Figure 4. Structure of cholesterol consensus motif (CCM). CCM in the structure of β2-adrenergic receptor (3D4S) as viewed with TM2 (A) or TM4 (B) in front. Orientation, extracellular side up. Principal residues of CCM are coloured. Green—Y70 in TM2, blue R151, yellow—W158 and pink—I154 in TM4.
Although CCM, CRAC, and CARC motifs appear in the sequence of large number GPCRs [17][42], CLR was found in structures lacking a CLR-binding motif, e.g., cannabinoid CB2 receptor (6PT0) or endothelin receptor (5X93) (Table 2). Binding of CLR to a detected CLR-binding motif was confirmed only in some of the published structures. For example, CCM was found at all five subtypes of the muscarinic acetylcholine receptor. However, no CLR was detected at any of the 16 published structures. In structures of the M1 receptor (5CXV and 6WJC), CHS is bound to CCM [43][44]. In many structures possessing a CLR-binding motif, CLR-binding was detected somewhere else. One of the abundant non-canonical CLR-binding sites is in the inner leaflet of the membrane at TM1 and Helix 8, e.g., structures 2RH1 (β2-adrenergic); 4IB4, 5TVN, and 6DRX (5-HT2B); and 6OS1 (AT1). The binding of CLR in this site is stabilized by palmitic acid covalently bound to the cysteine in Helix 8. Another non-canonical CLR-binding site appears in the outer leaflet of the membrane at TM6, e.g., structures 6PT2 (δ-opioid); 6B73 (κ-opioid); 4EIY and 5IU4 (A2A-adenosine). At structures 4DLK and 5C1M of the μ-opioid receptor, CLR binds to the variation of CCM. In these structures, the CLR hydroxyl group makes a hydrogen bond with Q314 instead of basic R or K of classic CCM. At structure 6LFM of the CXCR2 receptor, CLR binds to the variation of CRAC that possesses W instead of Y.
Table 2. Cholesterol binding motifs and residues interacting with CLR. List of X-ray structures of Class A GPCRs containing cholesterol predicted CLR-binding motifs and CLR-interacting residues. Code—PDB ID code, CLR—monomeric or dimeric state of CLR, leaflet—location of CLR in the inner or outer leaflet of the membrane, TM—transmembrane helices CLR is interacting with.
|
Rec. |
Code |
CLR |
Leaflet |
TM |
Predicted |
Confirmed? |
CLR-Interacting Residues |
|---|---|---|---|---|---|---|---|
|
α2C |
6KUW |
Monomer |
Out |
1, 7 |
CCM |
No |
Q45, Y46, E112, K420 |
|
β2 |
2RH1 |
Dimer |
In |
2, 3, 4 |
CCM |
Yes |
Y70, T73, S74, R151, W158 |
|
Monomer |
In |
1, 8 |
T56, C341_Plm |
||||
|
β2 |
3D4S |
Dimer |
In |
2, 3, 4 |
CCM |
Yes |
Y70, T73, S74, R151, W158 |
|
β2 |
6PS0 |
Monomer |
In |
2, 3, 4 |
CCM |
Yes |
Y70, T73, S74, R151, W158, F166 |
|
κOR |
6PT2 |
Monomer |
Out |
6 |
CCM |
No |
F280, D293 (o2) |
|
κOR |
6B73 |
Monomer |
Out |
6 |
CCM |
No |
T288, F293, T302, S303, S311 |
|
κOR |
6VI4 |
Monomer |
In |
4, 5 |
CCM |
No |
F147, T150, Y157, H162 |
|
μOR |
4DKL |
Monomer |
Out |
6 |
CCM Q |
Yes |
T294, Y299, F313, Q314, S317 |
|
μOR |
5C1M |
Monomer |
Out |
6 |
CCM Q |
Yes |
T294, Y299, F313, Q314, S317 |
|
5-HT2B |
4IB4 |
Monomer |
In |
1, 8 |
CCM |
No |
T73, Y394, Y399, C397_Plm |
|
5-HT2B |
5TVN |
Monomer |
In |
1, 8 |
CCM |
No |
T73, Y394, Y399 |
|
5-HT2B |
6DRX |
Monomer |
In |
1, 8 |
CCM |
No |
T73, Y394, Y399 |
|
A2A |
4EIY |
Dimer |
Out |
6 |
F182, 183, 255, 258, S263, H264 |
||
|
Monomer |
Out |
2, 3, 4 |
CCM |
No |
F70, F79, Q163 (o2) |
||
|
A2A |
5IU4 |
Dimer |
Out |
6 |
F182, 183, 255, 258, S263, H264 |
||
|
Dimer |
Out |
2, 3, 4 |
CCM |
No |
F70, H75, F79, F133, Q163 |
||
|
AT1 |
6OS1 |
Monomer |
In |
1, 8 |
CCM |
No |
F39, F44, S47 |
|
CB1 |
6N4B |
Monomer |
In |
3, 4 |
CRAC |
Partly |
F208, K232 |
|
Monomer |
In |
3, 4 |
F208, Y215, H219, R220 |
||||
|
CB2 |
6PT0 |
Dimer |
Out |
6 |
None |
-- |
Q276, K279, F283 |
|
Monomer |
In |
6 |
Y207, H211, W214, H217, R238, D240 |
||||
|
Monomer |
In |
3, 4 |
D130, Y137, T153, R149 |
||||
|
CCR9 |
5LWE |
Monomer |
In |
6 |
CCM |
Yes |
K254, T258, F263, F308, F319 |
|
CLT2 |
6RZ7 |
Monomer |
In |
5 |
CARC |
Yes |
S218, Y221, R226, F257 |
|
CXCR2 |
6LFM |
Monomer |
In |
2, 3, 4 |
CRAC W |
Yes |
N89, N129, K163, W170, L174 |
|
CXCR3 |
5WB2 |
Dimer |
Out |
6 |
CCM |
Yes |
T247, F265, S268, R271, T276 |
|
ETB |
5X93 |
Monomer |
Out |
1 |
None |
-- |
Y102, T105 |
|
FPR2 |
6LW5 |
Monomer |
In |
6 |
S215 |
||
|
Monomer |
In |
2, 3, 4 |
CRAC |
No |
N66, W150 |
||
|
FPR2 |
6OMM |
Dimer |
Out |
1, 2 |
-- |
F37, |
|
|
Monomer |
Out |
6 |
F206, F255, W267 |
||||
|
Monomer |
In |
6 |
H229 |
||||
|
Monomer |
In |
3, 4, 5 |
CRAC |
No |
F118, H129, W132 |
||
|
OTR |
6TPK |
Monomer |
Out |
4, 5 |
CCM |
No |
F191, W195, Y200, W203 |
|
P2Y12 |
4NTJ |
Monomer |
Out |
1, 7 |
F28, Y278, S282, W285 |
||
|
Monomer |
In |
3, 4 |
CCM |
No |
Y123, Q124 |
||
|
P2Y12 |
4PXZ |
Monomer |
In |
2, 3, 4 |
CCM |
Yes |
F51, S55, K64, N65, F106, K142, W149, F153 |
|
P2Y1 |
4XNV |
Monomer |
In |
2, 3, 4 |
CCM |
No |
Y189, T221, Y217, S213 |
Besides X-ray crystallography, approaches of computational chemistry also predicted interaction of CLR with GPCRs at many sites [4][45][46]. Multi-scale simulations of molecular dynamics revealed that CLR-interaction sites are dynamic in nature and are indicative of ‘high occupancy sites’ rather than ‘binding sites’. The results suggest that the energy landscape of CLR association with GPCRs corresponds to a series of shallow minima separated by low barriers. However, extensive all-atom simulations of molecular dynamics of the β2-adrenergic receptor (3D4S) suggest that CLR interacts specifically with the CCM and its binding is stable over the course of simulation [17]. CLR binding to the CCM of the β2-adrenergic receptor requires a slow, concerted rearrangement of side chains [47].
The structures of the A2A adenosine receptor 4EIY and 5UI4 contain three and four CLR molecules, respectively, that are bound at two nearly opposite positions at the extracellular side of the receptor (Figure 3A,C) [26][27]. However, none of them interacts with CCM detected at the intracellular half of TM2 and TM4. Simulation of molecular dynamics of the system containing the A2A-adenosine receptor in lipid bilayer containing 30 % of CLR resulted in the association of CLR with CCM and stabilization of TM6 by the CLR dimer [26][48]. The same approach identified additional CLR binding sites on the A2A receptor [49].
Two molecules of CLR were successfully docked to the site at the intracellular half of TM6 of the M1 muscarinic receptor (5CXV) identified by site-directed mutagenesis (Figure 5) [50]. Simulation of molecular dynamics of the docked CLR confirmed the stability of CLR binding and identified the hydrogen bond to R365 (R6.35 according to Ballesteros-Weinstein numbering [51]) as the key interaction.
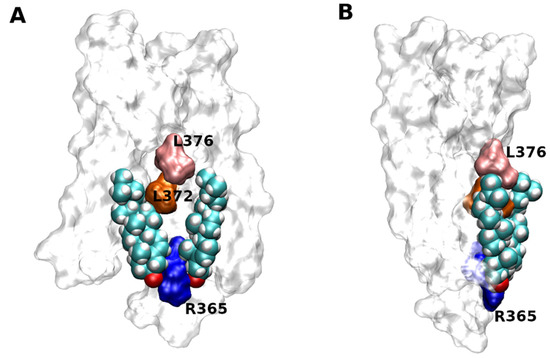
Figure 5. Docking of CLR to 5CXV structure of M1 muscarinic receptor. Two molecules of CLR docked to the structure of M1 muscarinic receptor (5CXV) as viewed with TM6 (A) or TM4 (B) in front. Orientation, extracellular side up. Principal residues in TM6 interacting with CLR are coloured. Blue—R365, orange—L372, pink—L376.