1000/1000
Hot
Most Recent

The plant hormone abscisic acid (ABA) triggers cellular tolerance responses to osmotic stress caused by drought and salinity. ABA controls the turgor pressure of guard cells in the plant epidermis, leading to stomatal closure to minimize water loss. However, stomatal apertures open to uptake CO2 for photosynthesis even under stress conditions. ABA modulates its signaling pathway via negative feedback regulation to maintain plant homeostasis. In the nuclei of guard cells, the clade A type 2C protein phosphatases (PP2Cs) counteract SnRK2 kinases by physical interaction, and thereby inhibit activation of the transcription factors that mediate ABA-responsive gene expression. Under osmotic stress conditions, PP2Cs bind to soluble ABA receptors to capture ABA and release active SnRK2s. Thus, PP2Cs function as a switch at the center of the ABA signaling network. ABA induces the expression of genes encoding repressors or activators of PP2C gene transcription. These regulators mediate the conversion of PP2C chromatins from a repressive to an active state for gene transcription. The stress-induced chromatin remodeling states of ABA-responsive genes could be memorized and transmitted to plant progeny; i.e., transgenerational epigenetic inheritance.
The current global climate crisis has resulted in long spells of dry weather and a shortage of rainfall, and becomes a serious threat to crop productivity and food supply. Under drought conditions, the salt concentration increases as the moisture content decreases in the soil. Water deficit and salinity inflict osmotic stress on plant cells. Plants are not able to escape from adverse environments, and so respond to such stressful conditions by triggering physiological and cellular responses[1][2][3]. Most prominently, plants close stomatal apertures on the epidermis to limit transpiration and thereby prevent loss of water under osmotic stress conditions. A stomatal aperture is formed by two flanking guard cells that swell or deflate by regulating turgor pressure through ionic fluxes via ion channels anchored in the plasma membrane[4].
Under osmotic stress conditions, plants biosynthesize and accumulate abscisic acid (ABA), a sesquiterpenoid hormone [5]. Most importantly, ABA functions as a chemical messenger that induces numerous genes whose products are crucial for stomatal closure and the accumulation of osmoprotectants[6][7][8]. A previous transcriptomic study showed that more than half of the genes regulated by ABA treatment are also induced under drought or salinity conditions[9]. Likewise, ABA deficiency impairs osmotic stress regulation of gene expression[10]. Thus, it appears that osmotic stress-induced expression of the responsive genes is entirely dependent on ABA. Because plants encounter not only osmotic stress but also abnormal temperatures (heat and cold) and biotic stresses (pathogens and insects) in nature, ABA signaling is integrated with other ABA-independent signaling pathways[11][12].
ABA is mainly biosynthesized in vascular tissues and transported to sites of action, such as guard cells[13][14]. In guard cells, ABA molecules are perceived by receptors in the nucleus and cytosol, activating the sucrose non-fermenting 1-related protein kinase 2 (SnRK2) family of protein kinases[15][16]. In the nucleus, SnRK2s phosphorylate a number of transcription factors that activate transcription of the ABA-responsive genes whose products are implicated in stress responses and tolerance. Inversely, the clade A type 2C protein phosphatases (PP2Cs) counteract SnRK2s by physical interaction, exerting negative regulation of ABA signaling[17]. Under osmotic stress conditions, PP2Cs bind to ABA receptors to capture ABA, releasing and activating the SnRK2s. Thus, PP2Cs function as a switch at the center of the ABA signaling network.
In Arabidopsis, nine protein phosphatases are classified as clade A PP2Cs[18][19][20]. Six of them—ABA insensitive 1 (ABI1), ABI2, ABA hypersensitive germination 1 (AHG1), AHG3/PP2CA, hypersensitive to ABA1 (HAB1), and HAB2—are involved in ABA signaling in the osmotic stress response. The remaining three members, highly ABA-induced 1 (HAI1), PP2C1/HAI2, and HAI3, affected ABA-independent low water potential phenotypes, such as enhanced accumulation of osmoprotectants and suppression of the expression of abiotic stress-associated genes encoding dehydrins and late embryogenesis abundant proteins (LEAs)[21]. ABI1 and ABI2 are main components of ABA signaling under abiotic stresses and in developmental processes[22][23]. The dominant ABA response mutants of Arabidopsis, abi1 and abi2, were originally isolated on the basis of their ABA insensitivity reflected in reduced seed dormancy and in symptoms of withering[24]. However, it was subsequently found that all of the knockout mutants of PP2C genes exhibited significant ABA hypersensitivity, indicating that they are negative regulators of ABA signaling. Recessive hab1-1 mutants also showed enhanced ABA-responsive gene expression, increased ABA-mediated stomatal closure, and ABA-hypersensitivity in seed germination, indicating that HAB1 also negatively regulates ABA signaling[25][26].
ABA also plays pivotal roles in various physiological processes during the plant life cycle, including seed dormancy, germination, lateral root formation, light signaling convergence, and control of flowering time[5][7][12]. These functions of ABA are related to Ca2+ influx, the production of reactive oxygen species such as H2O2, ion transport, and electrical signaling[11[12][27]. During these processes, ABA signaling interacts antagonistically or synergistically with other hormonal signaling pathways mediated by auxin, cytokinin, ethylene, and jasmonates[7]. Thus, excess ABA impairs developmental processes such as senescence, as well as pollen fertility, and also leads to seed dormancy and susceptibility to diseases[28].
High levels of PP2Cs are part of the negative feedback mechanism that desensitizes plants to high ABA levels[29][30]. In the absence of ABA, PP2Cs physically interact with SnRK2s to form complexes (Figure 1A). In Arabidopsis, subgroup III SnRK2s are key regulators of ABA signaling[31][32]. There are 10 SnRK2 members in Arabidopsis; i.e., SnRK2.1–SnRK2.10. Among them, SnRK2.2, SnRK2.3, and SnRK2.6/OST1 are the strongest activators of ABA responses, and so are regarded as primary regulators of ABA signaling. The triple mutation (snrk2.2/2.3/2.6) largely blocked the major ABA responses [33]. ABI1 interacts with SnRK2.6/OST1, SnRK2.2, and SnRK2.3 in plants, resulting in the inactivation of downstream components; e.g., AREB/ABFs transcription factors and ion channels[32].
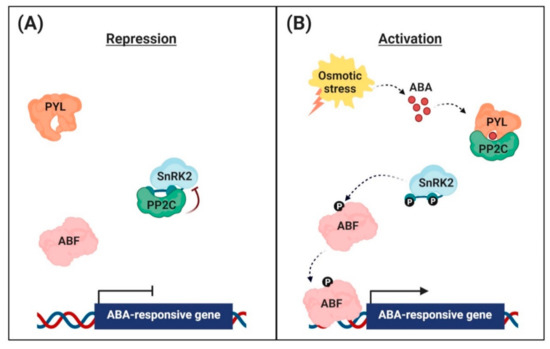
Figure 1. Abscisic acid (ABA) signaling pathway in the nuclei of guard cells. (A) Repression of ABA-responsive gene expression. In the absence, the clade A protein phosphatases (PP2Cs) physically interact with the sucrose non-fermenting 1-related protein kinases (SnRK2s) to reduce kinase activity via dephosphorylation. This inhibits the activity of ABRE-binding (AREB)/ABRE-binding factor (ABF) transcription factors and suppression of ABA-responsive gene transcription. (B) Activation of ABA-responsive gene expression. Under osmotic stress conditions, the interaction with ABA leads to conformational changes in the ABA receptors [PYR (pyrabactin resistance)/PYL (PYR-related)/RCAR (regulatory component of the ABA receptor)], allowing them to interact with PP2Cs. PP2Cs act as a coreceptor to capture ABA, thereby suppressing its phosphatase activity. This sequestrates PP2Cs from SnRK2s, and free SnRK2s phosphorylate the downstream transcription factors AREB/ABFs. The phosphorylated AREB/ABFs trigger the transcription of numerous ABA-responsive genes.
The SnRK2.6/OST1 was characterized as a critical limiting component in ABA regulation of stomatal apertures, ion channels, and NADPH oxidases in Arabidopsis guard cells[34]. PP2Cs dephosphorylate Ser175 in the activation loop of SnRK2.6, resulting in deactivation of the kinase [17]. Several PP2C-interacting factors, such as enhancer of ABA coreceptor 1 (EAR1) and PR5-like receptor kinase 2 (AtPR5K2), enhance the phosphatase activity of PP2Cs by phosphorylating them, and so modulate plant responses to drought stress[35][36].
ABA molecules biosynthesized in vascular tissues are distantly transmitted to sites such as guard cells to activate the closure of stomata[13][14]. Multiple ABA transporters have been identified in Arabidopsis, including exporters (AtABCG25 and AtDTX50) and importers (AtABCG40 and AtAIT1) [37][38][39][40][41]. Guard cells themselves also biosynthesize ABA, which is sufficient for stomatal closure in response to low air humidity[42] [56].
ABA molecules are perceived intracellularly by soluble receptors predominantly located in the nucleus and cytosol of guard cells [16][43][16,57]. A number of synonymous ABA receptors, e.g., pyrabactin resistance (PYR), PYR-related (PYL), and regulatory component of the ABA receptor (RCAR), have been identified as PP2C-interacting proteins in Arabidopsis[44][45][46]. PP2Cs have direct physical interactions with ABA and ABA receptors; these interactions are required for high-affinity binding of ABA [47][48]. Each PP2C functions as an ABA co-receptor within a holoreceptor complex that is constructed in combination with a particular PYR/PYL/RCAR.
The Arabidopsis genome contains 14 PYR/PYL/RCAR genes, which encode small proteins containing highly conserved amino acid residues[49]. All of them (except PYL13) are able to activate ABA-responsive gene expression. Transgenic lines expressing nuclear PYR1 in an ABA-insensitive mutant background exhibited ABA responses, but cytosolic PYR1 was also required for full recovery of ABA responses[50]. PYL8/RCAR3 showed subcellular localization mainly in the cytosol and nucleus, and its overexpression led to enhanced drought resistance of Arabidopsis[51]. Guard cells express the six ABA receptor genes PYR1, PYL1, PYL2, PYL4, PYL5, and PYL8 to mediate stomatal closure[52][53]. Arabidopsis mutants lacking three, four, five, and six of these PYR/PYL/RCAR genes (pyr1/pyl1/pyl2/pyl4/pyl5/pyl8) exhibited gradually increased stomatal conductance, indicating that this family of receptors quantitatively regulates the stomatal aperture [52]. Dittrich et al.[53] proposed that response specificity is achieved when the signals stimulate different members of the PYR/PYL/RCAR receptor family; PYL2 is sufficient for ABA-induced guard cell responses, whereas PYL4 and PYL5 are essential for the responses to CO2. Different combinations of PYRs and PP2Cs influence ABA binding affinity, and therefore affect the ABA sensitivity of the whole plant[54][55].
ABA directly binds to the PYR/PYL/RCAR proteins [47][48][56][57]. ABA binding leads to conformational changes of the ABA receptors, which allows physical interaction with PP2Cs and inhibits phosphatase activity [58][59][60] (Figure 1B). Nishimura et al.[60] performed co-immunoprecipitation experiments in a transgenic Arabidopsis line stably transformed with yellow fluorescent protein (YFP)–ABI1 fusion genes using a PYR1 antibody, and observed that the ABI1–PYR1 interaction was induced within 5 min after exogenous ABA application. Remarkable similarity was found in PP2C recognition between SnRK2 and ABA receptors [61][62]. In the absence of ABA, PP2C binds to the SnRK2 kinase domain and dephosphorylates Ser 175 in the activation loop. Upon perception of ABA, ABA receptor binds to PP2C by inserting the gate loop into the PP2C active cleft.
Upon the formation of PYL-ABA-PP2C complexes, SnRK2s dissociate from inactivated PP2Cs and recover their kinase activity. ABA treatment and osmotic stress stimulate phosphorylation of Ser 175 in the activation loop of SnRK2.6[63]. When released from PP2C inhibition, SnRK2.6 autophosphorylates at Ser175 and Thr176 to recover full activity[62]. Free and active SnRK2s subsequently phosphorylate and activate downstream transcription factors in the nucleus and ion channels in the cytosol[43].
In the nucleus, the SnRK2-mediated phosphorylation of transcription factors results in the expression of numerous ABA-responsive genes. By analyzing the promoters of ABA-responsive genes, a conserved ABA-responsive element (ABRE; PyACGTGG/TC) was identified[64][65]. Subsequently, a number of ABRE-binding (AREB) proteins and ABRE-binding factors (ABFs) were identified by yeast one-hybrid screenings[66][67]. AREB/ABFs belong to the basic-domain leucine zipper (bZIP) transcription factor family and are colocalized with SnRK2s in plant cell nuclei[32]. Multiple conserved RxxS/T sites in AREB/ABFs are phosphorylated in an ABA-dependent manner [67][68][69].
Among the nine AREB/ABFs in Arabidopsis, ABF1, AREB1/ABF2, ABF3, and AREB2/ABF4 act as master transcription factors in ABA signaling for osmotic stress tolerance[70]. Overexpression of these genes in Arabidopsis resulted in ABA hypersensitivity and enhanced drought stress tolerance[71][72][73]. By contrast, the triple knockout mutant (areb1/areb2/abf3) displayed impaired expression of ABA- and osmotic stress-responsive genes, resulting in increased sensitivity to drought[74]. Fujii et al. [75] reconstituted ABA-triggered phosphorylation of ABF2/AREB1 in vitro by combining PYR1, ABI1, and SnRK2.6/OST1, demonstrating that PYR/PYL/RCAR receptors, PP2Cs, and SnRK2s constitute the core of the ABA signaling pathway.