1000/1000
Hot
Most Recent

The Nrf2 is a transcription factor and it belongs to cap ‘n’ collar (CNC) basic leucine zipper (bZIP) protein subfamilies.
Under healthy conditions, there is a balance between antioxidant defense systems and free radical generation. However, under impaired balance between antioxidants and oxidants, oxidative stress develops, and cellular damage occurs [1], which is featured by multiple pathological chronic diseases, and the severity of viral infections like as recently pandemic of coronavirus disease 2019 (COVID-19) as a contagious respiratory and vascular disease [2]. Nrf2 (nuclear factor erythroid-derived 2-like 2 (NFE2L2)), as a stress-responsive transcription factor, effectively activates gene products and reduces reactive oxygen species (ROS) and electrophiles; thereby, the progression of various types of chronic diseases are prevented or retarded [3][4]. Keap1 rigorously regulated Nrf2, a key sensor of oxidative stress, and Cullin 3, in order to keep Nrf2 in the cytoplasm and actively present it for ubiquitination and proteasomal degradation [5]. Under oxidative stress, Keap1 releases Nrf2, which leads its translocation from cytosol to cellular nucleus, resulting in the expression of the antioxidant enzymes through an association with the antioxidant response element (ARE) and small musculoaponeurotic fibrosarcoma (sMaf) in order to combat oxidative stress and maintain cellular redox homeostasis [6][7]. Thereby, Nrf2 and associated proteins account as an ideal target against oxidative stress-related diseases, including neurodegenerative diseases, cancers, viral infection, cardiomyopathy, insulin resistance, and ischemia-reperfusion injury, as well as to protect other tissues or organs by regulating its level [8][9]. Nowadays, nutraceuticals and functional foods, including natural compounds, have been highlighted more than ever toward health keeping, disease prevention, and treatment [10]. Among them, curcumin (also called diferuloylmethane) is the main natural polyphenol of turmeric that has been traditionally used, due to its anti-inflammatory, anti-diabetic, anti-oxidant, anti-microbial, and anti-carcinogenic properties [11][12][13][14][15][16]. It has been shown that curcumin mediates the mitochondrial dysfunction inhibition and Nrf2 release/translocate into the nucleus in order to regulate the cell antioxidant pathways and provide cell survivability [9][17].
The Keap1–Nrf2 pathway plays a crucial role in protecting cells against endogenous and exogenous oxidative stresses and xenobiotic damages [18][19]. Kelch ECH associating protein 1 (Keap1) is the key component of the pathway. However, some accessory proteins also contribute to the cytoprotective function of the process, including ARE and sMaf proteins [20].
The Nrf2 is a transcription factor and it belongs to cap ‘n’ collar (CNC) basic leucine zipper (bZIP) protein subfamilies. The human Nrf2 comprises 605 amino acid residues, which is folded in seven highly conserved Nrf2-ECH (Neh1-7) domains [21][22]. Among them, the Neh1 domain contains a bZIP motif, which mediates dimerization and DNA binding while using leucine zipper (L-Zip) and a basic region structure, respectively. Neh2 plays a role in rapidly binding to Keap1 that is mediated by ETGE and DLG motifs (representing high and low binding affinities, respectively) and Keap1-dependent ubiquitination and degradation that are mediated by a region of seven lysine residues and the Keap1–Cul3 E3 ligase complex, and then this complex degrade by the 26S proteasome. Cullin 3 (Cul3) is a subunit of the E3 ligase complex that has a role as a scaffolding protein to keep close Rbx1 (RING-box protein 1)-Ub (ubiquitin)-loaded E2 and Keap1. Additionally, the 26S proteasome degrades Nrf2 by recognizing the phosphorylation of specific serine residues in the Neh6 domain of Nrf2. This process keeps the cellular Nrf2 at low levels [23][24]. Nrf2 has a rapid turnover and it presents a half-life of about 20–30 min. due to its constant degradation by the ubiquitin-proteasome system [25]. The Neh6 domain plays the role of a degron to mediate Nrf2 degradation in the nucleus mediating by phosphorylation of sites at specific serine residues [26]. Keap1–Nrf2 binding has been modeled as “hinge and latch”. According this model, the Kelch domain of the Keap1 at β- propeller conformation binds to the Neh2 domain of Nrf2 protein through high- and low-affinity bindings to the ETGE motif (as hinge) and DLG motifs (as latch), respectively [25].
Keap1 is a member of the BTB (bric-a-brac, tram-track, and broad-complex) Kelch family of proteins, which consists of 624 amino acid residues. Keap1, which is a zinc-metalloprotein with 27 cysteine residues, has five structural regions: N-terminal region (NTR), BTB domain, intervening region (IVR), Kelch domain, and C-terminal region (CTR). The BTB domain mediates the homodimerization of Keap1 and Cullin3 (Cul3) binding contains a sensor Cys151 residue. Additionally, IVR also contains a number of reactive cysteine residues, including Cys273 and Cys288. The Kelch domain with a six-bladed β -propeller conformation is responsible for Keap1 binding to the DLG and ETGE motifs in the Neh2 domain of Nrf2. Keap1 binds to Cul3 through both its BTB and IVR domains, targeting substrates for ubiquitination [23][27][28][29]. Cysteine residues, including Cys151, Cys273, and Cys288, are targets of electrophiles and oxidants, and their modification pattern leads to conformational changes in the Keap1, which disrupts the interaction between the Nrf2 that is mediated by Neh2 domain and Keap1 Kelch domains in order to remove the suppression of Nrf2 and then cancel its polyubiquitination [30]. Cys273 and Cys288 control basal and stress conditions of the cells, whereas, under cellular stress, Cys151 locates in the BTB domain is further reactive; likewise, specific toxins appear to modify Cys226, Cys434, and Cys613, thus disrupting the Keap1–Nrf2 complex [30][31]. The two cysteine residues, Cys273 and Cys288, in the central linker domain of Keap1, are involved in repressing Nrf2-dependent transcriptional activation [32]. Thereby, cysteine modification could be used as a therapeutic strategy for modulating the Keap1–Nrf2 complex. Natural compounds, such as curcumin, could be a potential drug to control the cell signaling pathway in stress conditions through Cys151 modification [23].
Under the normoxidant status of the cell, Keap1 keeps and suppresses Nrf2 in the cytosol in order to facilitate its ubiquitination by its E3 ligase activity in assembly with Cul3 and degradation in 26S proteasome. Meanwhile, under oxidative stress or electrophilic status, the modification of Keap1 at three candidate cysteine residues brings about altering the conformation of Keap1, which weakens Keap1–DLG, and, as a result, Nrf2 detachment and translocation to the nucleus [23]. In the nucleus, Nrf2 combines with the sMaf protein in order to form a heterodimer to activate ARE, which results in increasing the expression of a list of antioxidant enzymes [8]. The list includes heme oxygenase-1 (HO-1), NADP(H) quinone oxidoreductase 1 (NQO1), catalytic subunit (GCLC), glutamate-cysteine ligase modifier subunit (GCLM), CAT, SOD, thioredoxin reductase 1 (TrxR1), and aldo-keto reductase1 subunits C-1 (AKR1C1), C-2 (AKR1C2), and C-3 (AKR1C3) [3][8][33]. Moreover, plenty of functions are activated, including redox regulation, protein homeostasis, mitochondrial physiology, amino acid metabolism, iron metabolism, DNA repair, and some cellular processes, are prevented including apoptosis, autophagy, and proteasomal degradation [1][24].
The Keap1–Nrf2-ARE pathway shows a vital role in protecting cells from oxidative stress, which results in numerous human diseases. Targeting Nrf2 activation in the context of protein–protein interactions (PPIs) has emerged as an attractive strategy for designing innovative drugs to modulate Keap1–Nrf2 signaling and the transcription and expression of cytoprotective proteins, which is crucial in preventing oxidative stress-related pathological conditions. The Keap1–Nrf2 interaction inhibitors have been shown to be beneficial in many clinical indications. Direct disruptor or modifiers of the Keap1–Nrf2 complex could interrupt or induce conformational changes in the Keap1 structure. Later, Nrf2 is released and then moved from cytosol into the nucleus in order to activate the downstream antioxidant response element pathway. Accordingly, drug design-based Nrf2 activation strategies can be divided into modifying Keap1, leading to its conformational changes and disturbance of the Keap1–Nrf2. Numerous research studies described various Keap1–Nrf2 inhibitors, Keap1 modifiers, and Nrf2 activators [8][34].
Small molecules, like analogs of naphthalene-based or N-biphenyl substituted pyrazole carboxylate, have the potency to inhibit Keap1–Nrf2 interactions at the nanomolar scale, subsequently providing drug-like properties [35]. Dimethyl fumarate (DMF, Tecfidera), which is clinically used for the treatment of MS (Multiple sclerosis), is one of the modifiers of the Keap1–Nrf2 complex and it could covalently bind to the Cys151 residue of the Keap1–BTB domain. Bardoxolone (CDDO) reversibly reacts with the Cys151 residue of Keap-1, is designed to treat kidney disease in diabetic patients, but failed in phase 3 clinical trial due to the associated cardiac disease risks [8][27]. Although some of these small molecules are able to bind Keap1 at nanomolar potency, they exhibit moderate or even no inhibition activity against the Keap1–Nrf2 interaction. Hence, not every binding potency of Keap1 indicates Keap1–Nrf2 inhibition. There are several ETGE-like motifs in the Keap1 structure, and most of the inhibitors that bind to them cannot inhibit the interactions between Keap1–Nrf2 [36].
The proteasomal degradation of Nrf2 could be disrupted by some electrophiles that bind to Keap-1 sulfhydryl groups of the cysteine residues. Sulforaphane and tert-butyl hydroquinone (electrophilic substances) could react with the Cys151 and Cys171 of Keap1 and then change the ligase conformation in order to promote the escape of Nrf2 from degradation [37].
Peptide-based blockers have been designed to disrupt the Keap1–Nrf2 interactions, but low membrane permeability, poor bioavailability, instability, and high polarity are the major limitations of their application as a drug. [38][39]. Among designed peptide blockers, two egg-derived peptides, DKK and DDW, and the indole ring of tryptophan tetrapeptide W4 like cyclic motif-based peptide have been reported to disrupt Keap1–Nrf2 interaction, but additional validation is needed [36]. Fragment-based Kelch domain peptides that could interrupt the direct interaction of Keap1–Nrf2 is designed. These peptides bind to Keap1 with nanomolar affinity [40][41].
Natural products, like curcumin, contain enone and ketone groups as Michael acceptors, which makes it prone to react with the key cysteine thiolate residues in Keap1 (Figure 1a) [41]. Figure 1 shows the structure of the curcumin and some of its derivatives and formulations. According to the mass spectroscopy analysis-data curcumin modifies Cys151 of Keap1, resulting in conformation change to negatively impact the Keap1–Nrf2 PPI (Figure 2). Curcumin treatments of the cells with the mutations in Keap1–C273S, Keap1–C288S, and Keap-1-Cys151S could not activate Nrf2 translocation. Consequently, curcumin as an Nrf2 activator could be a potential drug to protect cells and although it has been considered as a natural therapeutic agent in various traditional medicines, but further clinical investigation is needed.
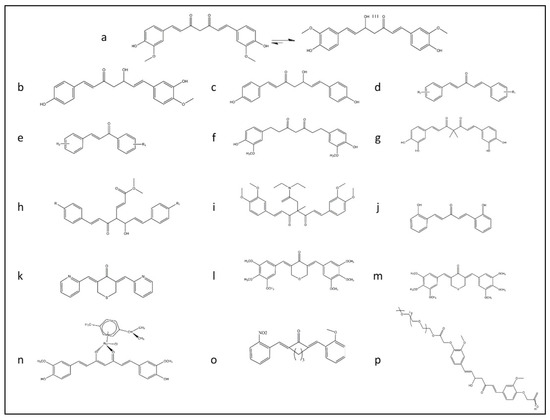
Figure 1. The rational design of derivative of curcumin for improving the bioavailability of this structure and promoting Nrf2 signaling pathway summarized in this figure. (a) Keto-enol form of curcumin structure. (b) Deoxymethylcurcumin. (c) Bisdeoxymethylcurcumin. (d) 5-carbon enone spacer analog of curcumin. (e) 3-carbon enone spacer analog of curcumin. (f) Tetrahydrocurcumin analog. (g) Curcumin analog with the geminal dimethyl groups and the catechol moiety. (h) Fumarate and curcumin-based analog. (i) TML-6 analog with modification for fix isomerization of keto form of curcumin with a methyl group and N, N-diethylacetamide. (j) Modified form of curcumin analog with five carbon linkages bis[2-hydroxybenzylidene] acetone (BHBA). (k) FN1 a synthetic analog of curcumin ((3E,5E)-3,5-bis(pyridin-2/3/4-methylene)-tetrahydrothiopyran-4-one). (l) E10 a synthetic analog of curcumin ((3E,5E)-3,5-bis-(3,4,5-trimethoxy-benzylidene)-tetrahydropyran-4-one). (m) F10 a synthetic analog of curcumin ((3E,5E)-3,5-bis-(3,4,5-trimethoxybenzylidene)-tetrahydro-thiopyran-4-one). (n) Ruthenium (II)-curcumin compound. (o) A13 a synthetic analog of curcumin (2,6-bis((3-methoxy-4-hydroxyphenyl) methylene)-cyclohexanone). (p) PEGylated analog of curcumin.
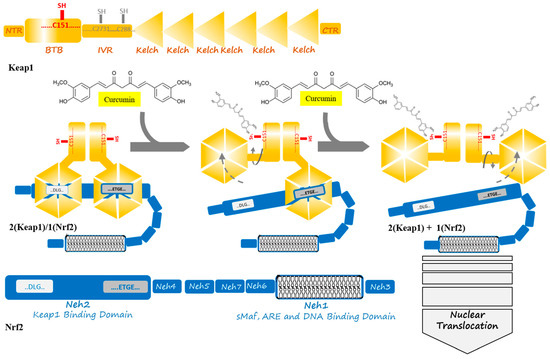
Figure 2. Curcumin induces a change in Keap1 conformation through binding to the sensor C151 residue of the Keap1 at the BTB domain. This binding is supposed to result in successive breaking of low- and high-affinity interactions between the Kelch domain of Keap1 and DLG- and ETGE motifs from Nrf2. The released Nrf2 is able to translocate to the cell nucleus to act as a transcription factor where starts anti-oxidant enzymes expression in an ARE and sMaf collaboration.
People around the world, for different remedies purposes, have consumed natural products. Turmeric is one of the most popular medicinal herbs with a long history of administration in India, Iran, and China. It has traditionally been applied to treat a broad range of diseases [42][43]. It is also commonly used in some Asian and African countries as a dietary spice. Curcumin is a hydrophobic polyphenol yellow pigment obtained from the dried rhizomes, of Curcuma longa, a rhizomatous herbaceous plant that belongs to the ginger family (Zingiberaceae) and it is a primary active compound of turmeric [11][12]. Curcuminoids complex, which is found in turmeric, consists of curcumin, demethoxycurcumin, and bisdemethoxycurcumin (Figure 1a–c, respectively) [13].
Extensive research shows that curcumin has a wide admiration range of beneficial properties, including anti-inflammatory, anti-diabetic, anti-oxidant, anti-microbial, anti-arthritic, anti-carcinogenic activity, and wound healing effects [14][15][16]. In vitro and in vivo studies demonstrated the ability of curcumin to modulate multiple cellular targets. Curcumin could modify multiple cell signaling pathways and downregulates the cell survival gene expression profile by its effects on transcription factors [44]. Curcumin affects the PI3K/Akt-1/mTOR, the Ras/Raf/MEK/ERK, the GSK-3β pathway, activates the p53 pathway, and regulates survival pathways via NF-κB, Akt, sand Nrf2/ARE pathways [45][46]. Curcumin exhibits antioxidant activity, because of donating hydrogen and scavenging free radicals. Besides, curcumin inhibits lipo-oxygenase (LOX) and cyclo-oxygenase (COX), xanthine oxygenase activities, nitric oxide synthesis, and ROS generation. Curcumin also inhibits the production of pro-inflammatory monocyte/macrophage-derived cytokines [interleukin- 8 (IL-8), monocyte inflammatory protein-1 (MIP-1), monocyte chemotactic protein-1 (MCP-1), interleukin-1b (IL-1b), and tumor necrosis factor-α (TNF-α) [47][48]. Consequently, curcumin is a non-toxic natural product, and its broad range of beneficial activities could be utilized as a therapeutic agent in various diseases.
Curcumin is able to suppress acute and chronic inflammation [11][47] and it has been asserted to penetrate the blood-brain barrier and moderate cerebral edema, reduce the inflammatory response, influence synaptic plasticity, and improve energy homeostasis through its direct and indirect antioxidant effects by eliminating ROS. Additionally, curcumin induces the expression of cytoprotective proteins through the Keap1–Nrf2 signaling pathway [49]. Besides, curcumin protects normal organs and sensitizes cancer cells through the activation of Nrf2. Nrf2 under activation by curcumin could inactivate the NF-κB and AP-1 signaling pathways, resulting in competitively blocking their activation by various stimuli [4][50]. Many recent studies have reported that oxidative stress, inflammation, and matrix degradation are essential contributors in arthritis, including temporomandibular joint (TMJ) osteoarthritis as a common stomatognathic disease. The curcumin treatment of human TMJ osteoarthritis effectively activates the Nrf2/ARE pathway in a dose-dependent manner [51]. The administration of curcumin significantly decreases H2O2 that is induced by oxidative damages and cell toxicity in osteoblast-like cells via retaining the glycogen synthase kinase 3 beta (GSK3β)-Nrf2 signaling pathway, which provides a possible promising osteoporosis treatment strategy [52]. The curcumin treatment of rat pheochromocytoma-derived cell line (PC12) exhibits a reduction of H2O2 that is induced by oxidative damages of DNA and increases cell viability under exposure to H2O2 [53]. Moreover, related studies to the current SARS-CoV-2 outbreak show that oxidative stress is associated with viral infection and pathogenesis. Therefore, the activation of antioxidant response genes and increased expression of heme oxygenase-1 (HO-1) and NAD(P)H:quinone oxidoreductase (NQO-1) through the Nrf2/ARE signaling pathway makes it a possible therapy strategy [54][55].