Fungi help maintain the diversity of the ecosystem and are critical to nutrient-cycling by degrading dead organic materials
[1]. In nature, fungi with agricultural, ecological, economic, biotechnological, and medical importance have been identified. On the other hand, fungi can also cause human infections. Invasive fungal diseases account for a majority of complications among immunocompromised patients worldwide
[2]. Fungi of the order Mucorales cause mucormycosis, a rare but highly fatal fungal infection. They can cause cutaneous, rhino-orbital, pulmonary, rhino-cerebral, and disseminated bloodstream infections; the severity and prognosis largely depend upon the infection type. The fatality rate is very high
[3] with 96% among patients with disseminated infections
[4]. The incidence of mucormycosis is rapidly increasing, especially in developing countries like India and Nepal, where Mucorales were mostly found to cause rhino-orbito-cerebral infections. The most recent incidences are observed in COVID-19 patients
[5][6][7] or those who recently recovered from COVID-19. Given that the incidence of mucormycosis has been associated with preexisting conditions such as uncontrolled diabetes mellitus, malignancies, trauma, or extended corticosteroids use
[4], it is likely that the number of cases with mucormycosis will increase further due to the current COVID-19 pandemic. Primarily found in soil and decaying vegetation, Mucorales are ubiquitous, have reduced susceptibility to most clinically used antifungal agents, and thrive under high acid conditions. Their thermotolerance, however, is greatly varying; some, such as
Rhizopus microspores thrive at temperatures as high as 50 °C
[8], while some, such as
R. sexualis cannot grow above 25 °C
[8]. Based on their thermotolerance, it was previously thought that Mucorales that cannot grow at 37 °C were medically not important. However, case reports and incidences of cutaneous infections caused by
M. hiemalis [9][10][11][12], which cannot grow at 37 °C
[13][14], have been observed. These cases of infections suggested that the fungi that do not grow at 37 °C are capable of causing superficial infections. Furthermore, invasive mucormycosis rapidly disseminates within the host tissues. Depending upon the infection site, Mucorales interact with specific host receptors and take advantage of the host physiological conditions to derive nutrients such as iron
[15]. Furthermore, Mucorales can store iron in ferritins besides siderophores, further depleting the host of iron and proliferating within the host rapidly
[16]. Therefore, patients with diabetes mellitus and COVID-19 are more prone to mucormycosis as these patients have a higher level of free iron in their blood
[17][18]. As mucormycosis has been increasingly common, studies on the understanding of their pathogenesis as well as identifying emerging virulent strains are of top priority.
2. Taxonomy and Phylogenetic Analysis
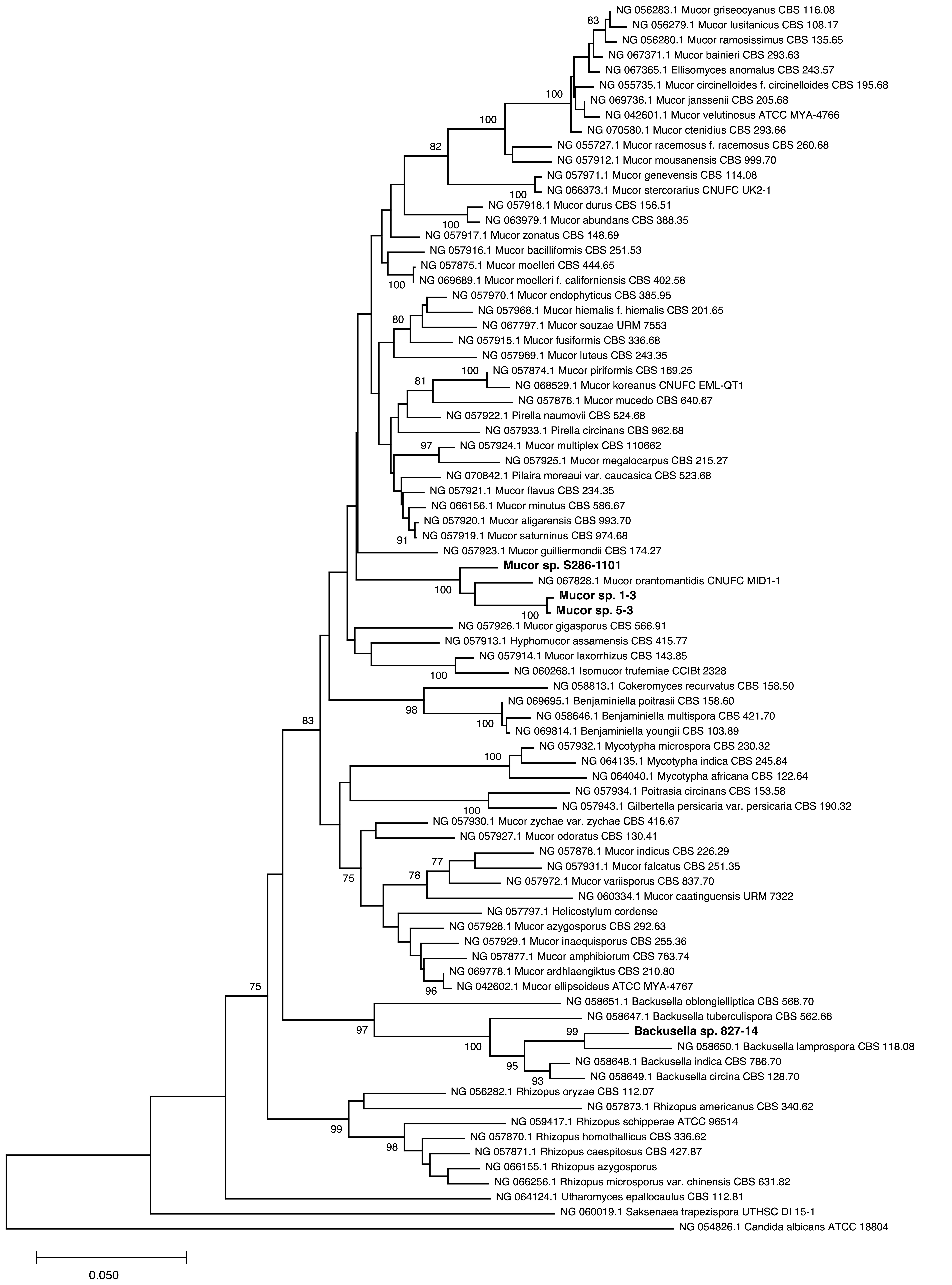
We isolated four fungal strains from plant sources and attempted to identify them (
Table 1). BLAST analysis of the LSU rRNA region showed that strains 1-3, 5-3, and S286-1101 were related to
Mucor orantomantidis (accession no: NG_067828.1) with the percentage identity of 95.4% (641/672 bp), 95.4% (641/672 bp), and 97.2% (651/670 bp), respectively. Strain 827-14 was 96% (624/650) identical to
Backusella lamprospora CBS 118.08 (accession no: NG_058650.1). This result was consistent with the constructed phylogenetic tree using MEGA X
[19], where we found that strains 1-3, 5-3, and S286-1101 were claded with
M. orantomantidis, and strain 827-14 was claded with
B. lamprospora (
Figure 1). These findings suggested that these strains were novel species belonging to
Mucor (1-3, 5-3, and S286-1101) and
Backusella (827-14). The taxonomic assignment of these strains was further confirmed by the phylogenic tree using ITS sequences, another locus used frequently for the taxonomic demarcation of Mucorales
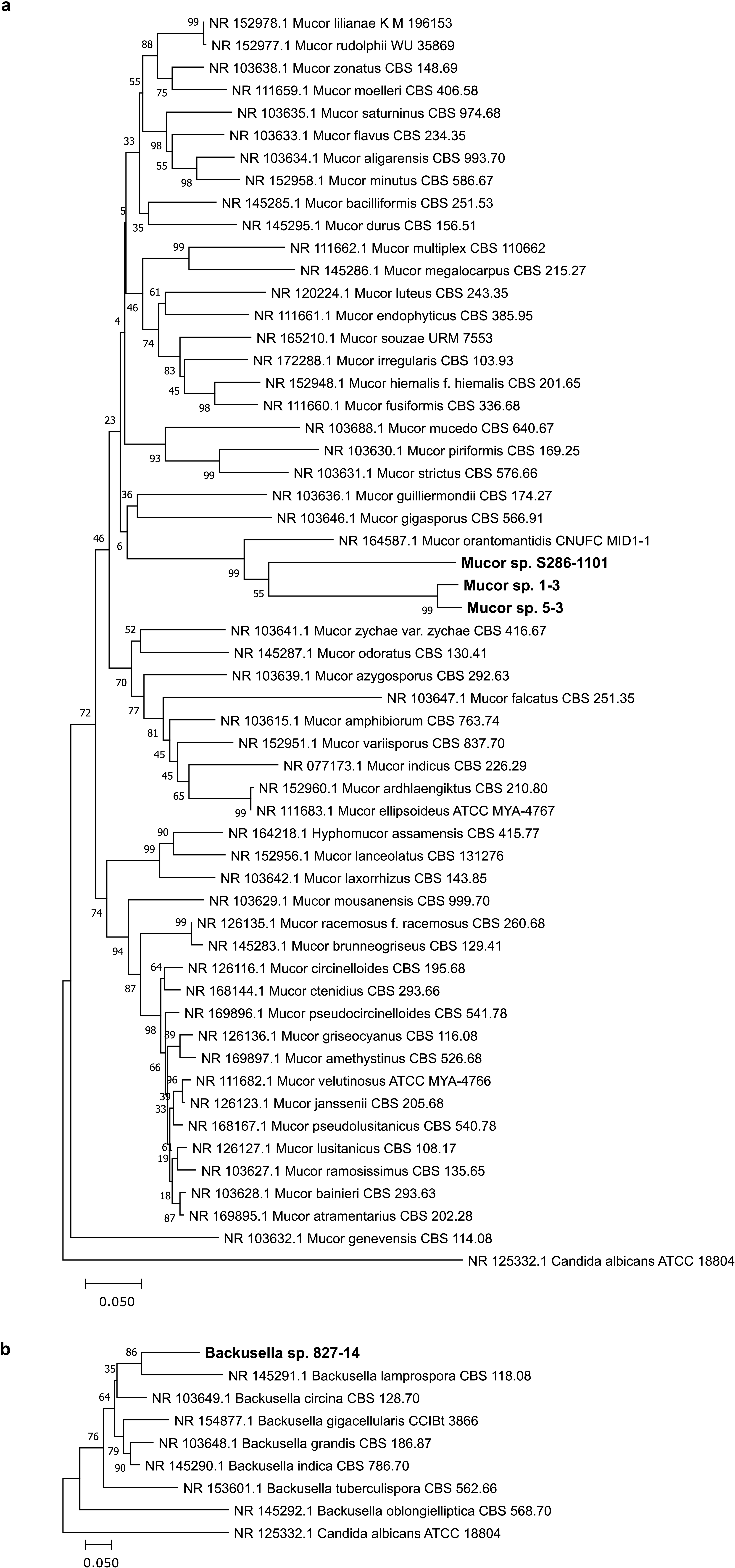
[13]. ITS analysis showed that
Mucor strains were claded with
M. orantomantidis (
Figure 2a) and
Backusella sp. 827-14 claded with
B. lamprospora (
Figure 2b). The phylogenetic tree was reconstructed using MAFFT
[20] and RAxML
[21] to reveal similar results. Among four strains, we randomly selected
Mucor sp. 5-3 for whole genome sequencing and SEM analysis.
Figure 1. LSU rRNA-based phylogenetic analysis of Mucorales. The LSU rRNA sequence was utilized to infer the evolutionary relationship of the strains. The optimal tree, drawn to scale, with branch lengths in the same units as those of evolutionary distances, with the sum of branch length = 2.44139183, is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. After the ambiguous positions were removed for each sequence pair, there were a total of 1552 positions in the final dataset. The novel strains are written in boldface with larger font size.
Figure 2. ITS-based phylogenetic analysis of Mucorales. The ITS sequence was utilized to infer the evolutionary relationship of the strains. The optimal tree, drawn to scale, with branch lengths in the same units as those of the evolutionary distances, with the sum of branch lengths of (a) 4.16205042 and (b) 1.37992789 for Mucor and Backusella, respectively, is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. After the ambiguous positions were removed for each sequence pair, there were a total of 1064 (a) and 975 (b) positions in the final dataset. The novel strains are written in boldface with larger font size.
Table 1. Source and the growth of novel fungi at different temperatures.
| Strain |
Source (Location) |
Growth at |
| 37 °C (4 Days) |
30 °C (4 Days) |
4 °C (10 Days) |
| Mucor sp. 1-3 |
Plant bud (Chiba, Japan) |
− |
+++ |
− |
| Mucor sp. 5-3 |
Plant seed (Chiba, Japan) |
+ |
+++ |
− |
| Mucor sp. S286-1101 |
Plant leaf (Saitama, Japan) |
+ |
+++ |
− |
| Backusella sp. 827-14 |
Dead plant leaf (Chiba, Japan) |
− |
+++ |
− |
3. Analysis of the Mucor Genome Assembly
We have previously used the next-generation sequencing tool for the genomic analysis of various bacteria
[22][23] and fungi
[24] and have found that compared to the bacterial genome, the fungal genome contains a large number of repeats and is difficult to assemble. We sequenced the
Mucor sp. 5-3 genome using the Ion PGM System and found that its genome size was 30.8 Mb in size, had a G+C content of 39.26%, and divided into about 17,079 contigs (
Table 2). This indicated a presence of a large number of repeat elements in the genome. Of note, the same sequencing approach, used for
Candida albicans TIMM1768 resulted in about 3400 contigs for a 14.5 Mb genome
[24]. Besides, combined with a previous study
[25], it can be expected that the
Mucor genome is diverse with a genome size ranging between 30–47 Mb.
Table 2. General features of Mucor sp. 5-3 draft genome.
| Features |
Characteristics |
| Total reads |
5,016,222 |
| Average length (bp) |
270 |
| Coverage |
44× |
| Genome size (bp) |
30,813,178 |
| Contigs |
17,079 |
| Contigs > 1000 bp |
6792 |
| Longest contig (bp) |
37,105 |
| N50 |
4436 |
| L50 |
2075 |
| G + C (%) |
39.26 |
4. Morphological Studies
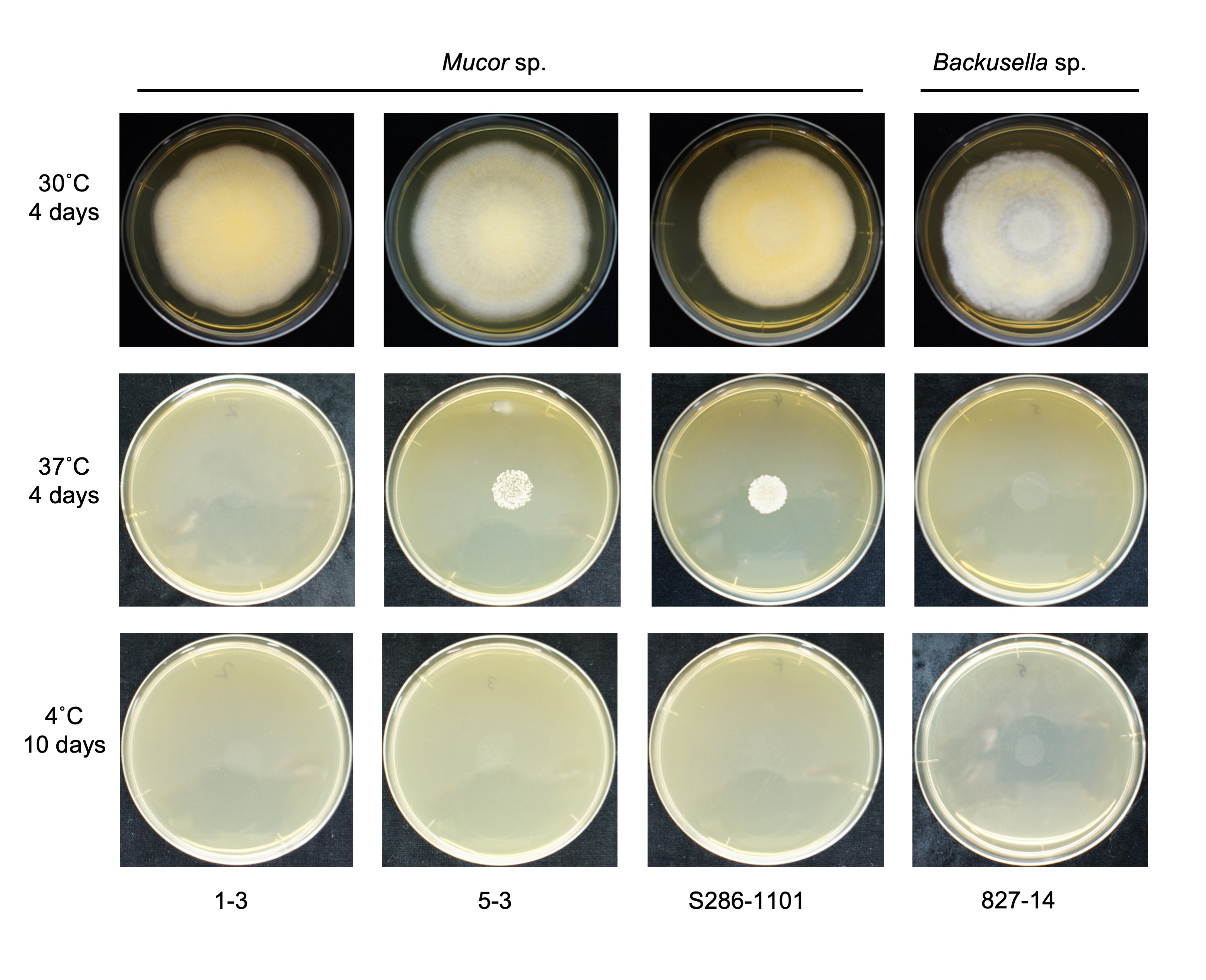
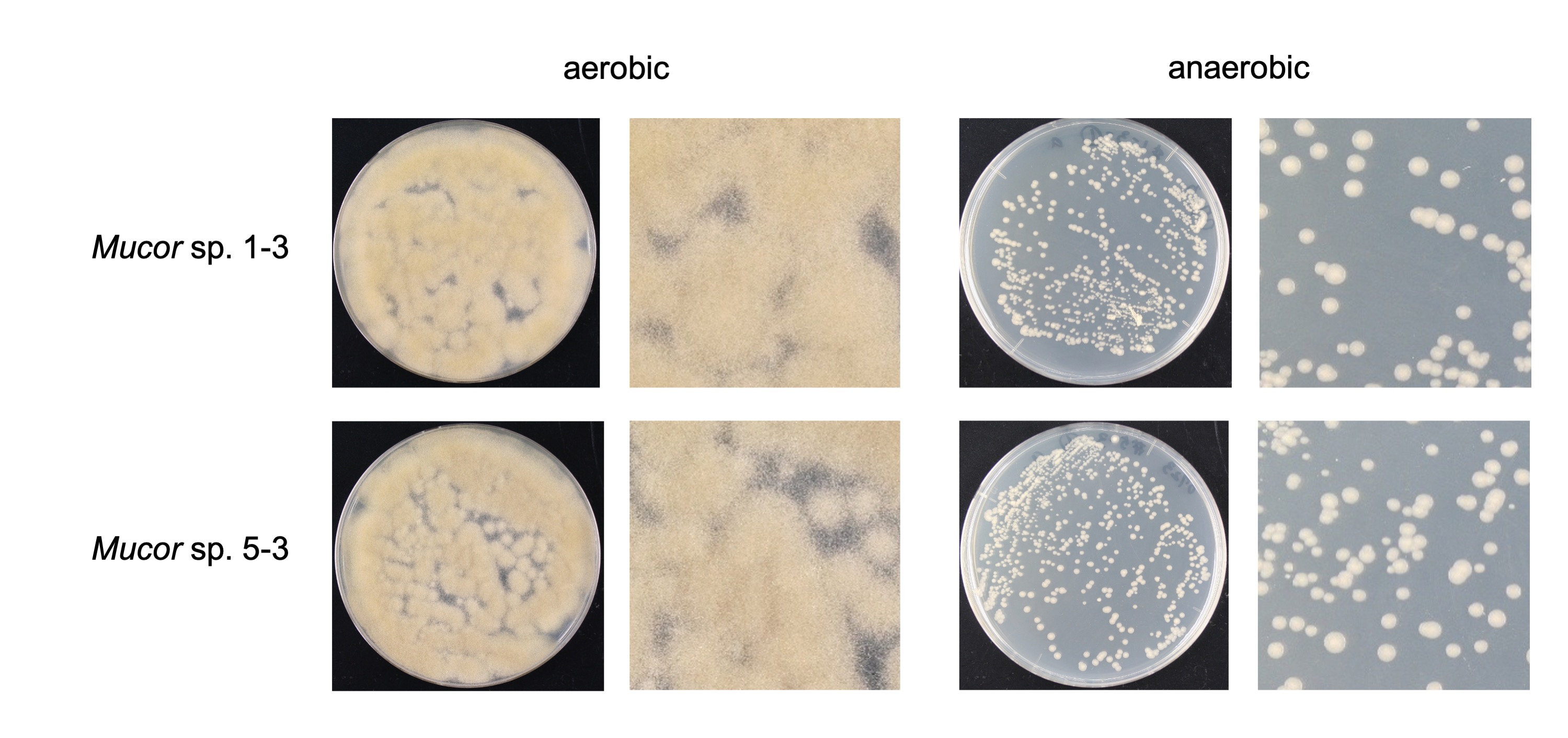
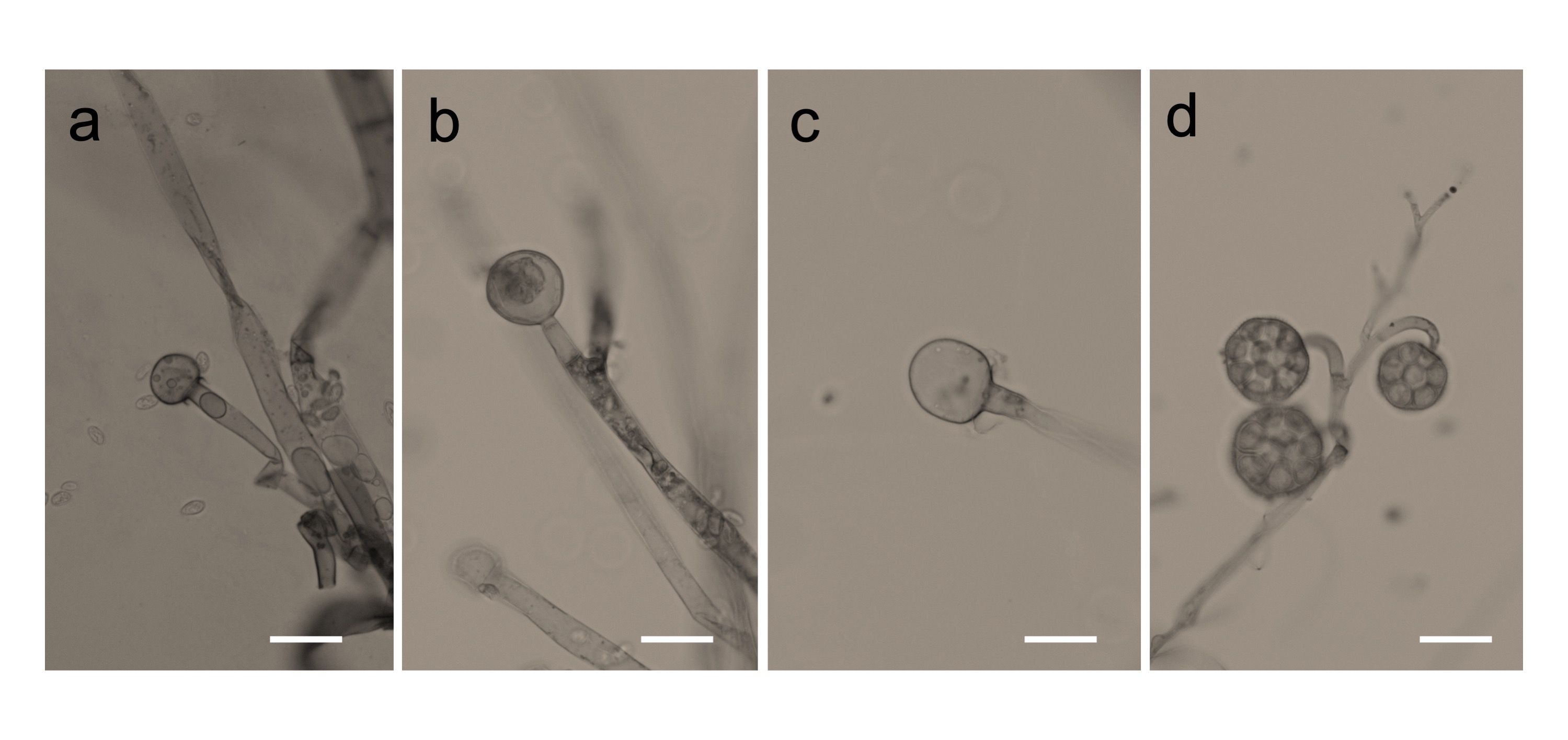
We found that all the strains grew well on the YPD agar plate at 30 °C when grown under aerobic conditions. However, they had a difference in their growth status at 4 °C and 37 °C. Mucor sp. 5-3, and S286-1101 could grow, although slowly, while the other two strains could not grow at 37 °C. At 4 °C, Mucor spp. did not show a visible sign of growth; however, Backusella showed a faint growth (Figure 3). Next, we used Mucor sp. 1-3 and Mucor sp. 5-3 to check their ability to grow under anaerobic conditions. Incubation for two days at 30 °C after streaking on YPD agar showed that the cells grew as yeast under anaerobic conditions; however, mycelial growth was observed under aerobic conditions (Figure 4). Besides, we found that the colony morphology—when visualized with the naked eye—of these two strains on agar plates was similar. When sporangia were observed under a light microscope, a difference was observed among Mucor spp. and Backusella spp. (Figure 5a–d).
Figure 3. Temperature-specific growth of novel Mucorales. 4.0 × 104 spores contained in 40 µL of normal saline were spotted at the center of the YPD agar plates, dried, and incubated at different temperatures for the designated duration before taking a picture.
Figure 4. Aerobic and anaerobic growth of Mucor sp. 1-3 and 5-3.
Figure 5. Photomicrographs of Mucorales grown on slide culture. Mucor sp. 1-3 (a), 5-3 (b), S286-1101 (c), and Backusella sp. 827-14 (d). Mucorales were grown aerobically for 4 days. Bars = 20 µm.
5. Mucor Morphology under High Resolution
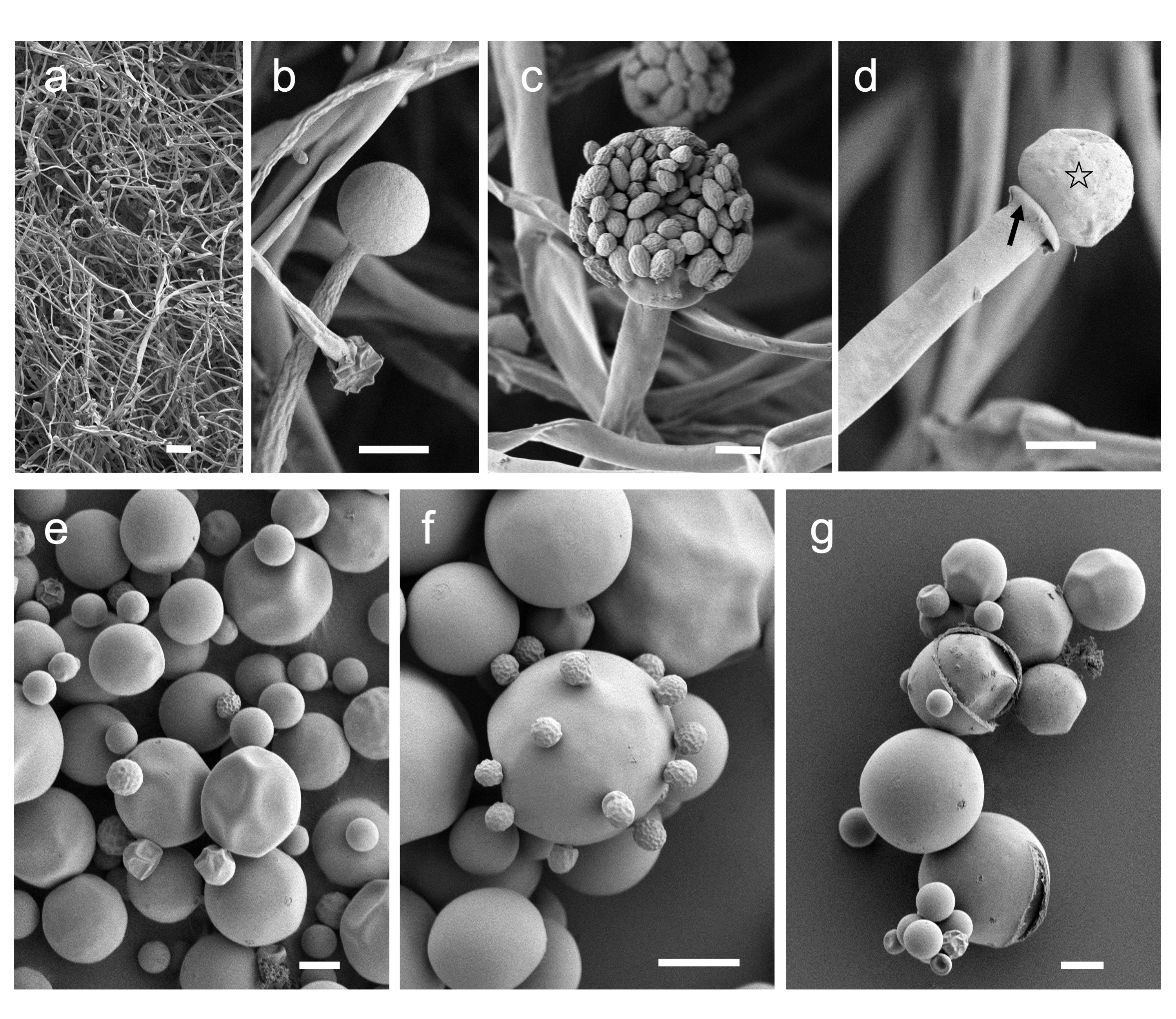
Using Mucor sp. 5-3, cell morphology of mycelial growth under aerobic conditions and yeast growth under anaerobic conditions were studied by Scanning Electron Microscope (SEM). Aerobic culture resulted in hyphal growth, where long elongated aseptate hyphae and sporangium were observed (Figure 6a,b). Inside the matured sporangium, a large number of sporangiospores were observed (Figure 6c). The tip of sporangiophore harbored columella and collarette (Figure 6d). The yeast colonies formed under anaerobic growth were made up of spherical cells of various sizes ranging from 2 to 30 µm in diameter (Figure 6e). Multipolar budding cells were observed in large cells (Figure 6e,f). Some budding cells had smooth surfaces, whereas some had small golf ball-like shapes (Figure 6f). Furthermore, strange cells with cleaved cell surfaces were often observed in large cells (Figure 6g). The existence of cells with cleaved surface layers has never been reported before. The significance of these cells in the growth of yeast cells needs to be investigated in more detail in the future.
Figure 6. SEM images of aerobic (a–d) and anaerobic (e–g) culture of Mucor sp. 5-3. (a) Low magnification image of mycelium; aseptate hyphae and sporangia. (b) Sporangia formed at the tip of the sporangiophore. (c) Sporangiospores inside the sporangia. (d) Columella (open star) and collarette (filled arrow) were observed at the tip of the sporangiophore. (e) Yeast-like colonies are composed of spherical cells of various sizes. (f) Multipolar budding found in large cells. (g) Cleavage of surface structure is observed in large cells. Mucor sp. 5-3 was grown aerobically (a–d) and anaerobically (e–g) on a YPD agar plate for 2 days. Bars: (a) = 100 µm, (b–g) = 10 µm.
6. Pathogenicity of Newly Isolated Mucorales
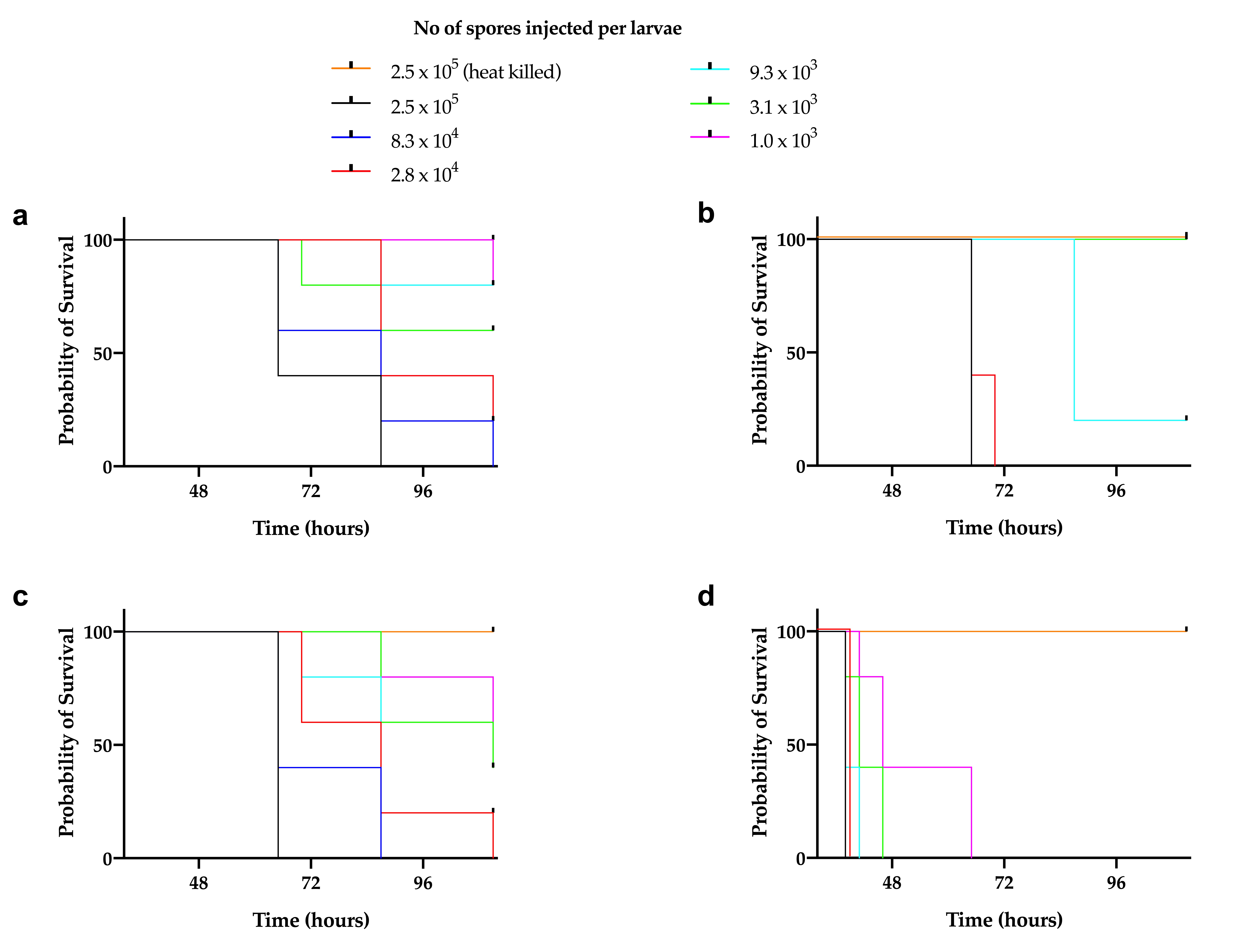
Although various vertebrates and invertebrates are used as animal models to study mucormycosis
[26], silkworms were not used to test the pathogenicity of mucoralean fungi so far. Given that the use of the silkworm infection model for the test of pathogenicity of several bacteria and fungi is well established
[27][28][29][30], we aimed to evaluate the pathogenicity of novel Mucorales using a silkworm infection model. First, we prepared a suspension of spores and examined the pathogenicity of the suspension without dilution. We found that all silkworms died at 20 h. Next, we prepared a 1/8—fold dilution of spore suspension, injected it into the silkworm, and checked the survival at 15 and 20 h. We found that at 15 h, most of the silkworms were surviving; however, the lethality of these Mucorales was high at 20 h, indicating the ability of these fungi to rapidly kill silkworms. These results indicated the pathogenicity of the spores of newly identified fungi against silkworms. Next, for a quantitative evaluation of pathogenicity, we serially diluted the spores and injected them into the silkworm hemolymph so that each silkworm received 2.5 × 10
5–1.0 × 10
3 spores. To confirm the establishment of infection, silkworms were inoculated with 2.5 × 10
5 heat-killed spores of each strain. Whereas the heat-killed spores were nonpathogenic to silkworms, a dose-dependent killing by live spores was observed (
Figure 7a–d). We found that with the same dose injected,
Backusella were more pathogenic compared to
Mucor.
Figure 7. Pathogenicity of Mucorales spores in silkworm. Fifty microliters serially diluted spore suspensions of Mucor sp. 1-3 (a), Mucor sp. 5-3 (b), Mucor sp. S286-1101 (c), and Backusella sp. 827-14 (d) were injected into the silkworm hemolymph (n = 5). Silkworms were incubated at 27 °C and survival was recorded at various time points.
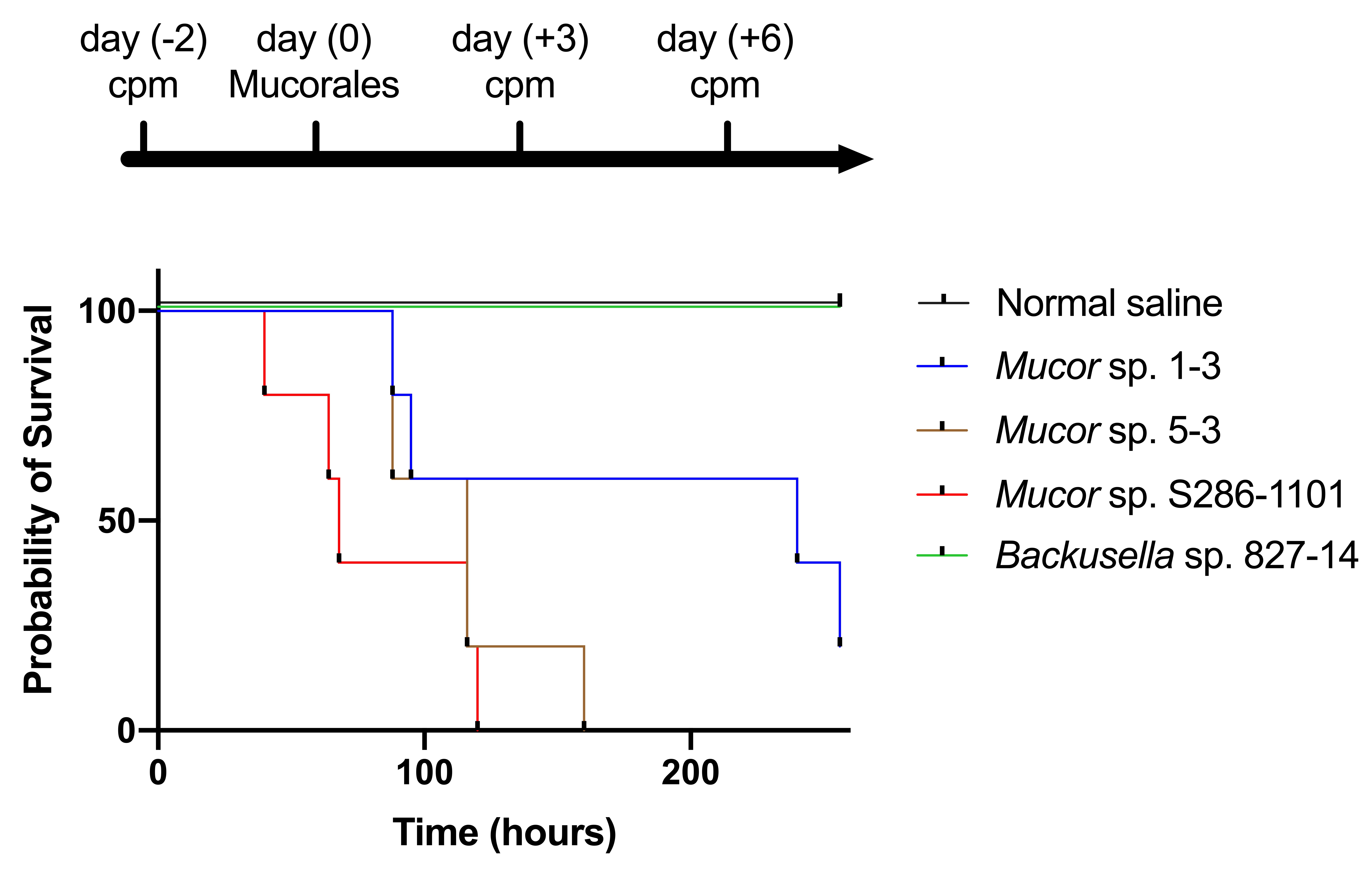
Next, we tested the ability of these strains to infect mammalian hosts using an immunocompromised mouse model. The result showed that three out of four strains could kill the mouse within four days of the infection suggesting their pathogenicity to mammals (Figure 8).
Figure 8. Pathogenicity of Mucorales spores in mice. Four weeks old ICR female mice (n = 5) were immunocompromised by injecting cyclophosphamide (cpm) and injected with 2.0 × 108 colony-forming units of Mucorales spores through the intraperitoneal route and survival was recorded at various time points.