1000/1000
Hot
Most Recent

Sterol regulatory element-binding protein-1c (SREBP-1c) is a key modulatory molecule involved in lipid homeostasis in the central nervous system. However, little is known about the biological effects of SREBP-1c in the brain. Our previous study uncovered that mice deficient in SREBP-1c exhibit schizophrenia-like behaviors. We analyzed the transcriptomes of the hippocampus of SREBP-1c knockout (KO) mice and wild-type mice in order to investigate whether there are novel molecular mechanisms involved in the neurological aberrations caused by SREBP-1c deficiency. This study found seven differentially expressed genes (three up-regulated and four down-regulated genes) in the hippocampus of SREBP-1c KO mice. For further verification, we selected the three most significantly changed genes: glucagon-like peptide 2 receptors (GLP2R) involved in hippocampal neurogenesis and neuroplasticity as well as in cognitive impairments; necdin (NDN) which is related to neuronal death and neurodevelopmental disorders; and Erb-B2 receptor tyrosine kinase 4 (ERBB4) which is a receptor for schizophrenia-linked protein, neuregulin-1. The protein levels of GLP2R and NDN were considerably decreased, but the level of ERBB4 was significantly increased in the hippocampus of SREBP-1c KO mice. We suggest that these data provide novel molecular evidence for the modulatory role of SREBP-1c in the mouse hippocampus.
SREBPs are a family of transcription factors that regulate lipid homeostasis by controlling the synthesis of cholesterol, fatty acids (FA), triacylglycerol, and phospholipids[1]. The SREBP family includes three isoforms: SREBP-1a, SREBP-1c, and SREBP-2[2]. SREBP-2 is encoded by the sterol regulatory element-binding transcription factor 2 (Srebf2) gene while both SREBP-1a and -1c are encoded by the Srebf1 gene as it has independent promoters that utilize alternate first exons that are then spliced into common exons[1][3]. SREBP-1c primarily regulates FA and triglyceride synthesis, SREBP-2 controls cholesterol synthesis, and SREBP-1a participants in the roles of both SREBP-1c and SREBP-2[4]. When the cellular lipid level is low, the SREBP cleavage activating protein transfers SREBP precursors from the endoplasmic reticulum to the Golgi apparatus, in which two distinct proteases sequentially cleave the SREBP precursors, which results in the release of the active forms[5][6]. The active forms subsequently translocate to the nucleus and regulate the transcription of target genes by binding to enhancer (E)-boxes and sterol regulatory element deoxyribose nucleic acid (DNA) sequences[7].
As SREBPs primarily play a role in lipid homeostasis by controlling the production of fatty acids and cholesterol, they have been widely studied in the liver and adipose tissue[8]. Lipid homeostasis plays an important role in neuronal function especially in the regulation of the membrane’s function between the intracellular and extracellular spaces, modulation of synaptic outputs, and release of lipid-derived second messengers[8][9]. Accumulating evidence has revealed that a disturbance in lipid homeostasis can cause several brain diseases, such as Huntington’s disease[10], Alzheimer’s disease[11], Parkinson’s disease[12], and mood disorders[9]. This confirms the importance of adequate SREBP function in the brain. Among the SREBP family isoforms, SREBP-1c is the predominant isoform that is considered to play a central role in lipid homeostasis[13][14]. Recently, we found that the deficiency of SREBP-1c induces schizophrenia-like behavior in mice[15], and further explore the mechanisms involved in these behavioral aberrations, we sought to identify differentially expressed genes (DEGs) in the hippocampus of SREBP-1c knockout (KO) mice compared to in that of wild type (WT) littermates.
Our main conclusions outline that the presence of alterations in the expression of novel gene candidates in the hippocampus of SREBP-1c KO mice is closely related to the development of schizophrenia-like behavior; further, we have identified the differential gene expression of various genes, which are potentially responsible for changes in signaling transduction, cell differentiation, and cell survival within the hippocampal cell population.
To identify DEGs in the SREBP-1c KO mice in comparison to in the WT mice, the EdgeR package was chosen since, up to 2018, it has been the most widely used by researchers[16] and has demonstrated better performance in identifying true positives[17] compared to Cuffdiff2 and DESeq. The ExactTest function in the EdgeR package was used to analyze DEGs[18]. We detected seven genes by using the false discovery rate (FDR) < 0.05 criteria in an RNA-seq dataset (Figure 1A). The divergence between the gene expression of the WT and SREBP-1c KO mice was confirmed by hierarchical clustering (Figure 1B). The results of a gene ontology analysis using the list of seven DEGs suggest that the signaling pathways, cell/neuron migrations, cellular responses, transcription, cell motility, and localization of cells are affected in SREBP-1c KO mice (Figure 1C) because the Srebf gene is suggested to act as a transcription factor and signal transduction regulator that controls lipid metabolism and insulin signaling[2].
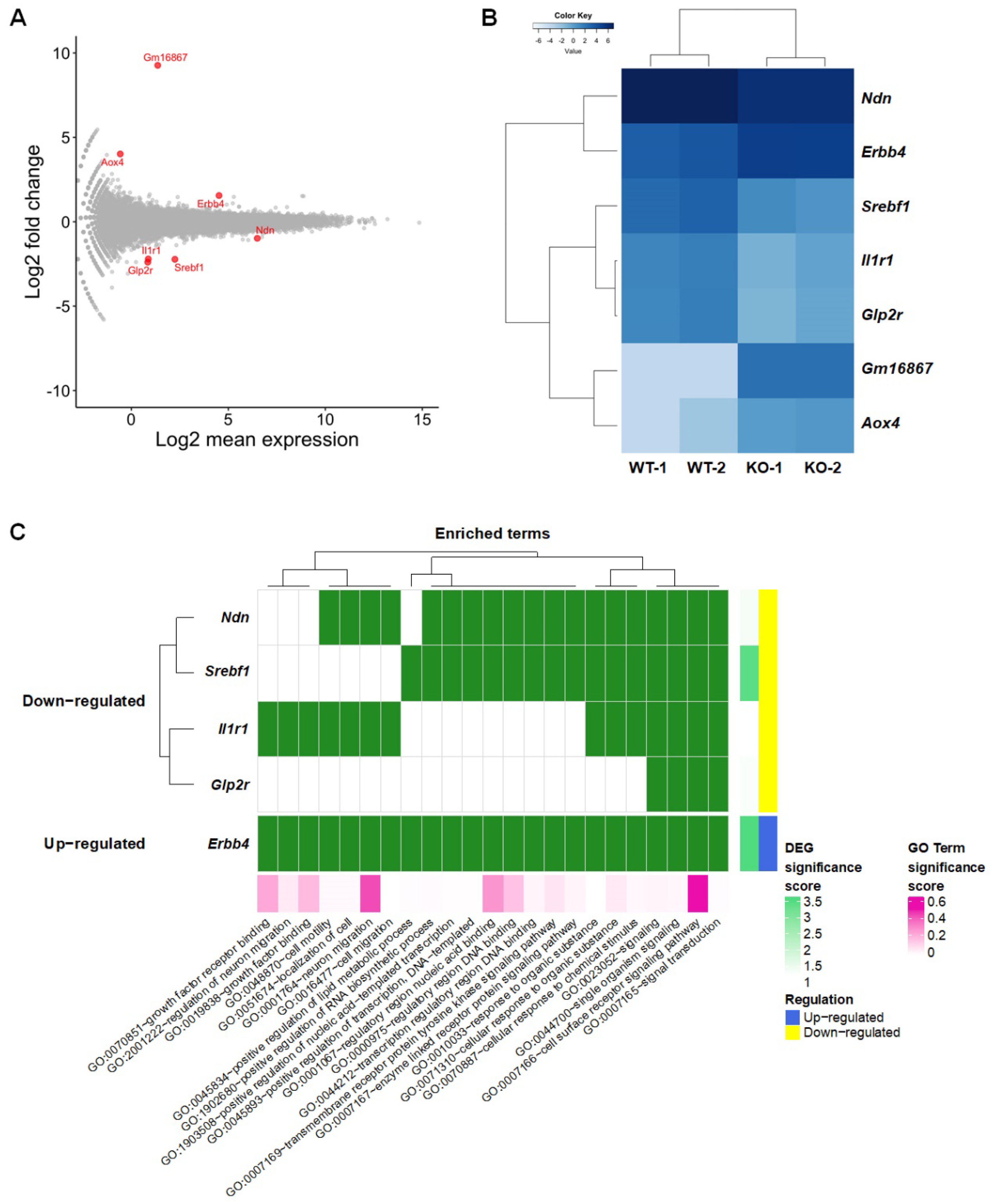
Figure 1. Distinct gene expression changes in the hippocampus of SREBP-1c KO mice. (A) MA plot and (B) Heat map showing DEGs (FDR < 0.05, |Log2 fold change| ≥1). (C) Gene ontology enrichment analysis. DEG significance score is expressed as −log2 (FDR). Aox4, aldehyde oxidase 4; DEG, differentially expressed genes; Erbb4, Erb-B2 receptor tyrosine kinase 4; FDR, false discovery rate; Glp2r, glucagon-like peptide 2 receptor; Il1r1, interleukin 1 receptor, type I; Ndn, necdin; Srebf1, sterol regulatory element-binding transcription factor 1.
Additionally, using the Search Tool for the Retrieval of Interacting Genes (STRING) database, among the DEGs identified only Erb-B2 receptor tyrosine kinase 4 (Erbb4) and interleukin 1 receptor type I (Il1r1) were found to interact with Srebfs[19]. Erbb4 had predicted interactions with Srebf1 predicted by co-citation analysis with pre-existing scientific texts and gene fusion analysis. For example, a previous study implicated Erbb4 as an activator of Srebf2 mediating increases in low-density lipoprotein uptake and cholesterol biosynthesis in mammary epithelial cells[5]. Il1r1, on the other hand, had predicted interactions with Srebf1 evidenced by text-mining, gene fusion analysis, gene co-expression, and a known interaction based on curated interaction records (Unpublished data).
To determine if the gene expressions detected via the qRT-PCR were similar to those detected via the RNA-seq analyses, we analyzed selected genes based on gene expression and biological relevance (Figure 2). A gene expression validation technique revealed that in the hippocampus of the SREBP-1c KO mice, the expressions of the glucagon-like peptide 2 receptor (Glp2r) (M = 0.09292, SD = 0.03947; t(14) = 14.38, p < 0.0001), necdin (Ndn) (M = 0.5874, SD = 0.1639; t(14) = 4.559, p = 0.0004), and Il1r1 (M = 0.4722, SD = 0.1774; t(14) = 2.835, p = 0.0132) genes significantly decreased (Figure 2A–C), while the levels of Gm16867 (M = 1.965, SD = 0.7913; t(14) = 3.124, p = 0.0075), Erbb4 (M = 1.188, SD = 0.2059; t(14) = 2.159, p = 0.0487), and aldehyde oxidase 4 (Aox4) (M = 2.323, SD = 1.724; t(14) = 2.148, p = 0.0497) were significantly increased (Figure 2D–F).
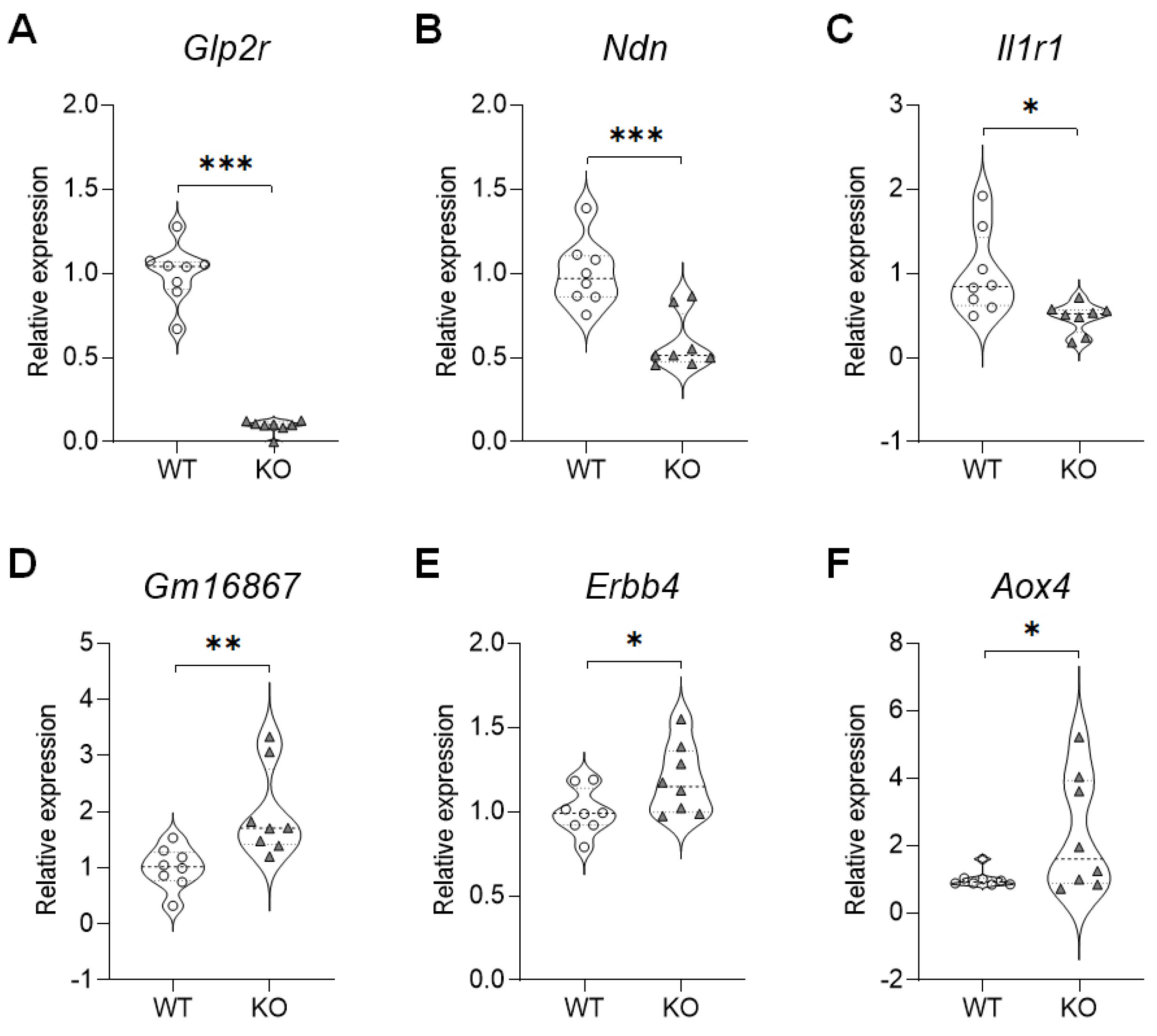
Figure 2. qRT-PCR validation of selected DEGs in the hippocampus of SREBP-1c KO mice. (A) Gip2r, (B) Ndn, (C) Il1r1, (D) Gm16867, (E) Erbb4, (F) Aox4 (n = 8, *p < 0.05, **p < 0.01 and ***p < 0.001 compared to WT mice). Aox4, aldehyde oxidase 4; DEG, differentially expressed genes; Erbb4, Erb-B2 receptor tyrosine kinase 4; Glp2r, glucagon-like peptide 2 receptor; Il1r1, interleukin 1 receptor type I; KO, SREBP-1c knockout mice; Ndn, necdin; qRT-PCR, quantitative real-time polymerase chain reaction; WT, wild-type mice.
To determine if the protein expression pattern that we identified corresponded with the results of the qRT-PCR analyses, western blot analyses were performed for selected differentially expressed genes, Glp2r, Ndn, and Erbb4 (Figure 3A). The results revealed that the total protein expression that was uncovered for decreased levels of GLP2R (M = 0.4345, SD = 0.1226; t(4) = 7.545, p = 0.0017) and NDN (M = 0.5140, SD = 0.08201; t(4) = 6.419, p = 0.0030), and increased level of ERBB4 (M = 1.935, SD = 0.3769; t(4) = 3.699, p = 0.0209) in the hippocampus was in agreement with the results of the RNA-seq analysis and qRT-PCR. Furthermore, these findings were validated by the immunohistochemical results that revealed similar alterations in the expression patterns of GLP2R (Figure 3B), NDN (Figure 3C), and ERBB4 (Figure 3D) in the hippocampus. In particular, GLP2R and NDN were strongly expressed in the PCL of the hippocampal CA3 subregion, which corresponds with the localizations of intense SREBP expressions found in this subregion. However, ERBB4 was expressed extensively in all subregions of the hippocampus.
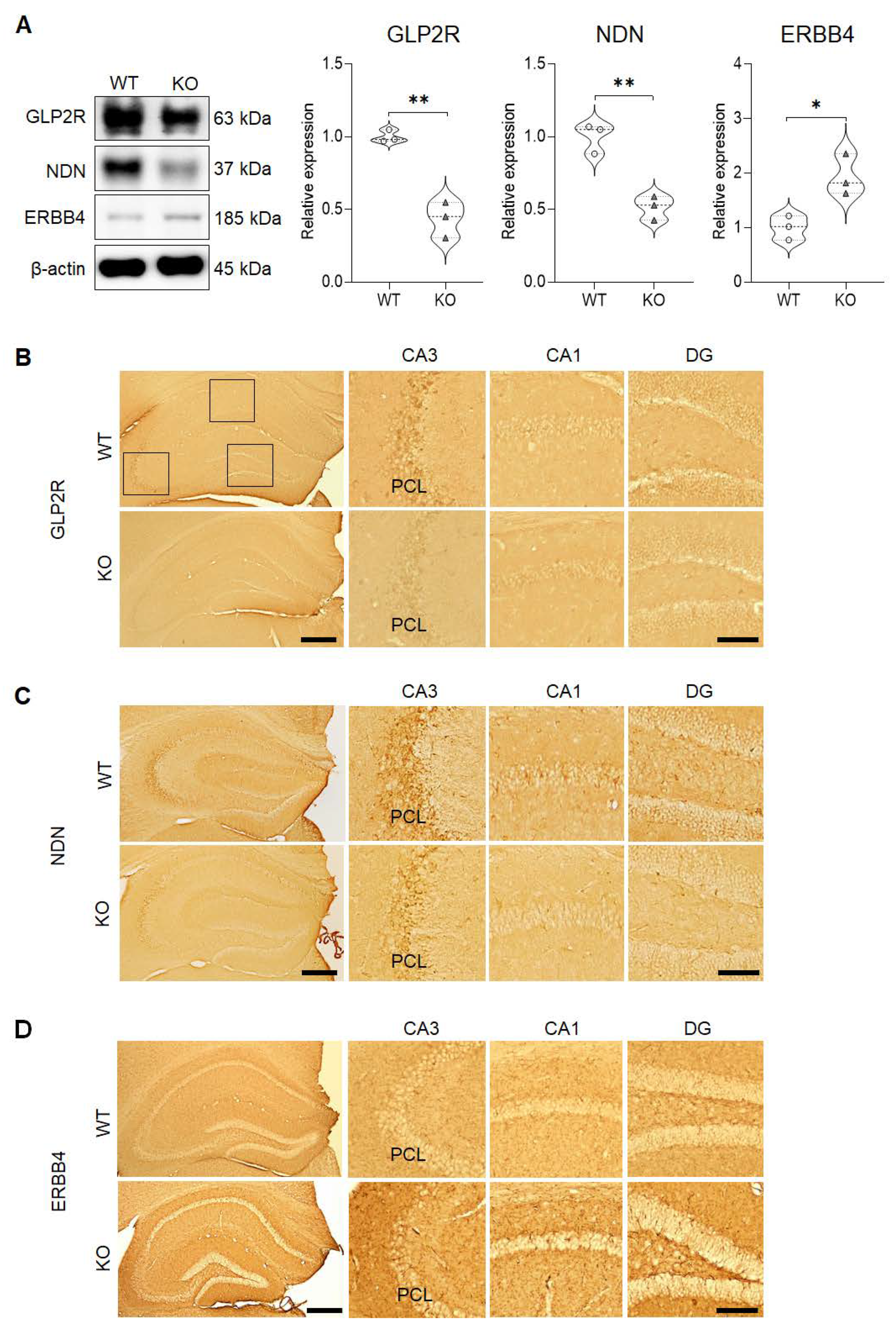
Figure 3. Validation of GLP2R, NDN, and ERBB4 protein expressions in the hippocampus. (A) Western blot analyses confirm that both GLP2R and NDN expressions are significantly decreased, while ERBB4 expression is significantly increased in the hippocampus of SREBP-1c KO mice compared to in those of WT mice (n = 3, *p < 0.05 and **p < 0.01 compared to WT mice). (B–D) Immunohistochemical analyses reveal that both GLP2R (B) and NDN (C) are expressed in the CA1, CA3, and DG subregions of the hippocampus (with especially intense immunoreactivities in the PCL of the CA3 subregion), while ERBB4 (D) is expressed extensively in all subregions of the hippocampus. Scale bars = 100 µm (left panels), 50 µm (Insets). CA, cornu ammonis; DG, dentate gyrus; ERBB4, Erb-B2 receptor tyrosine kinase 4; GLP2R, glucagon-like peptide 2 receptor; KO, SREBP-1c knockout mice; NDN, necdin; PCL, pyramidal cell layer; WT, wild-type mice.
Experimentally, we were able to validate our main results by performing various analyses that generated similar results, and therefore, in conclusion, based on the aforementioned findings, we can speculate that the aberrations in the behavioral patterns of the KO mice are potentially related to the differential gene expression of the seven genes that we identified. Thus, this study further provides evidence for the possible relationship between SREBP-1c and schizophrenia; it is the first study to reveal other molecules that are altered in conjunction with the depletion of SREBP-1c signaling in the mouse hippocampus.
Based on our results, we propose that the findings in the schizophrenia-like behavioral phenotype of SREBP1-c KO mice[15] are closely linked to the differential gene expression of Glp2r, Ndn, Il1r1, Erbb4, and Aox4, which possibly results in changes in signaling transduction, cell differentiation, and cell survival within the hippocampal cell population. However, further scrutiny of the translational value from rodent models to human patients needs to be done to fully establish the relationship with the differentially expressed genes found in this study and the conjectured hippocampal functional alterations. Furthermore, due to the multifaceted hypotheses surrounding the etiology of schizophrenia, it would be worthwhile to explore the developmental transcriptome system of this model in future studies. This would provide an avenue for the elucidation of possible DEGs related to the neurodevelopmental hypothesis of schizophrenia’s etiology. Nevertheless, acquiring this information represents an important starting point for the establishment of new models for schizophrenia and provides a novel avenue for future studies that may explore new therapeutic targets to treat schizophrenia.