The maintenance of genome integrity is critical for cell survival. Homologous recombination (HR) is considered the major error-free repair pathway in combatting endogenously generated double-stranded lesions in DNA. Nevertheless, a number of alternative repair pathways have been described as protectors of genome stability, especially in HR-deficient cells. One of the factors that appears to have a role in many of these pathways is human RAD52, a DNA repair protein that was previously considered to be dispensable due to a lack of an observable phenotype in knock-out mice. In later studies, RAD52 deficiency has been shown to be synthetically lethal with defects in BRCA genes, making RAD52 an attractive therapeutic target, particularly in the context of BRCA-deficient tumors.
1. Double-Strand Breaks and RAD52
Double-strand breaks (DSBs) are the greatest threat to genomic integrity and can result from endogenous sources such as reactive oxygen species (ROS) and replication stress or exogenous insults such as UV, ionizing radiation (IR), and chemotherapeutic compounds. If left unrepaired, DSBs can cause chromosomal aberrations, mutagenesis, and cell death, leading to the development of severe diseases including cancer
[1][2]. Given the cytotoxic nature of these lesions, the cell has developed multiple pathways of DSB repair, some which result in mutagenic repair, and others that use a template to ensure error-free repair.
The majority of DSBs in a cell are repaired by non-homologous end-joining (NHEJ), which ligates blunt ends in a sequence-independent manner and can result in small insertions or deletions (indels)
[1]. Homologous recombination (HR) is generally considered the most faithful DSB repair pathway, as it uses the intact sister chromatid as a donor, and it is highly dependent on breast cancer associated genes BRCA1 and BRCA2. The usage of a sister chromatid means that HR is predominately active in S or G2 when the cell has replicated its DNA
[3][4]. In order to initiate HR, the DSB is resected, leaving a segment of single-stranded DNA (ssDNA) that is coated first by RPA, then by RAD51 in order to facilitate homology search and invasion of the sister chromatid—a process known as strand exchange. BRCA1 is predominately involved in the resection step of HR, while BRCA2 mediates removal of RPA and loading of RAD51; however, recent work has shown that BRCA1 may also have a function in the later steps of HR
[3][5][6].
While NHEJ is the predominant pathway for DSB repair, its requirement for blunt ends renders it incapable of dealing with resected ends or one-ended DSBs resulting from replication fork collapse. Replication stress is the most common source of endogenous damage and is usually repaired by HR proteins
[7][8]. In an HR-deficient context, DSBs that cannot be repaired by NHEJ must be channeled through error-prone homology-mediated pathways such as single-strand annealing (SSA) and alternative end-joining (Alt-EJ). In SSA, long-range resection proceeds bi-directionally from the DSB until homologous sequences are revealed. The break is then repaired by the annealing of these complementary sequences, resulting in deletion of the intervening sequence
[3][9]. Alt-EJ operates by an analogous mechanism, only it requires a shorter length of resection and anneals regions of microhomology, resulting in smaller deletions. While the pattern of resection and annealing of complementary sequences are similar, Alt-EJ and SSA have been shown to require different protein factors
[3][9][10]. SSA has canonically been considered the only RAD52-dependent pathway, as it is responsible for annealing the resected ends
[9][11][12][13][14]; however, recent work by the Hendrickson lab using previously established plasmid reporter assays has shown that HR, Alt-EJ, and SSA all decrease in a RAD52-null cell
[15].
Rad52 (Radiation sensitive 52) was first identified in budding yeast in a screen for mutations that would inhibit repair of IR-induced DSBs, resulting in radio-sensitization
[16][17]. Several other DNA repair proteins were also identified in this screen and are referred to as the Rad52 epistasis group. Of this group, most proteins were found to play similar roles in yeast and human cells, while Rad52 had a marked difference between its functionality in yeast and in higher eukaryotes
[14].
While mammalian cells rely on BRCA2 to mediate loading of RAD51 onto RPA-coated ssDNA and on RAD52 for annealing ssDNA ends, in yeast, both of these functions are performed by Rad52, as they lack a functional homolog of BRCA2
[18]. Consequently, Rad52-mutant yeast show defects in meiotic recombination, spore viability, HR, and mating-type switching
[16], while RAD52-null mice display no observable phenotype in viability or fertility and only a mild effect on HR
[19]. Despite the apparent dispensability of RAD52 in mammals, one of the initial hypotheses for a function of RAD52 in human cells came from our lab’s work showing that BRCA2-deficient cells required RAD52 for survival, indicating a synthetically lethal relationship
[20]. In a BRCA-deficient context, RAD51 loading can be carried out by RAD52, albeit at a lower efficiency
[20][21]. A recent study in vitro has supported this function of RAD52 by using DNA curtain analysis to show the dynamics of interactions between RPA, RAD52, RAD51, and ssDNA
[22]. RAD52′s potential involvement in HR was also supported by the finding that GFP-tagged RAD52 formed foci in response to DNA damage throughout S and G2, coinciding with HR activity during these stages of the cell cycle
[23]. Since then, RAD52 has been implicated in many other pathways of DNA repair and genome maintenance, most of which are also dominant in S and G2 (
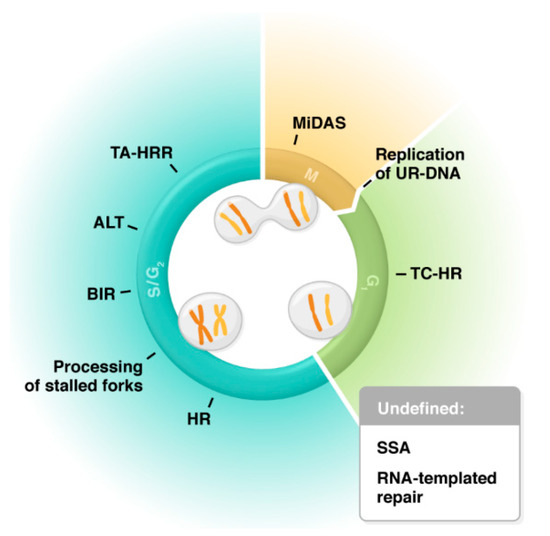
Figure 1)
[24][25][26][27][28][29][30].
Figure 1. Involvement of RAD52 in various pathways of genome maintenance: TC-HR (transcription-coupled homologous recombination), HR (homologous recombination), Processing of stalled forks (MUS81-dependent fork cleavage and protection against excessive fork reversal), BIR (break-induced replication), ALT (alternative lengthening of telomeres), TA-HRR (transcription-associated homologous recombination repair), MiDAS (mitotic DNA synthesis), Replication of UR-DNA (replication of under-replicated DNA in 53BP1 nuclear bodies), SSA (single-strand annealing), and RNA-templated repair. Pathways are categorized by the cell cycle phase in which they are predominately active
[24][25][26][27][28][29][30].
2. Biochemistry and Activity of RAD52
Human RAD52 consists of a C-terminal domain (CTD) which contains its nuclear localization signal (NLS) as well as its RAD51- and RPA-binding regions, and an N-terminal domain (NTD) which has been shown to be responsible for its multimerization and DNA-binding activity
[16][31][32][33][34][35]. RAD52 can also undergo context-dependent post-translational modifications such as SUMOylation, phosphorylation, acetylation, and methylation, the functions of which are under active investigation
[16][36][37][38][39][40].
Full-length RAD52 exists as a heptameric ring, while purified NTDs have been shown to form an undecameric ring structure
[41][42]. While a ring structure would usually be suggestive of a helicase, the DNA-binding domain of RAD52 is a groove of positively-charged residues which wraps around the outside of the ring, unlike helicases, which typically thread DNA through the center of the ring
[32][41][43]. This domain has been shown to bind DNA with a footprint of 4 nucleotides per monomer, meaning that each heptameric ring could bind around 28 bp of DNA
[43][44][45]. RAD52 has also been shown to contain a secondary DNA-binding site that can interact with ssDNA, which would facilitate annealing of resected ends, as in SSA. This site can also bind double-stranded DNA (dsDNA), which enables strand exchange in an ATP-independent manner
[31][46]. Recent studies using atomic force spectroscopy have provided insights into the mechanics of RAD52/DNA interactions and illuminated possible molecular mechanisms for RAD52-facilitated annealing and strand exchange
[47]. In addition to canonical or “forward” strand exchange, where RAD52 binds ssDNA and facilitates its invasion of dsDNA, RAD52 can also perform inverse strand exchange by binding the duplex DNA, which is then invaded by ssDNA or RNA
[48][49].
3. RAD52 as a Target for Cancer Therapy
Despite its importance in a variety of repair pathways, RAD52 has long appeared dispensable for cell viability except in the context of BRCA-deficiency. This makes it an attractive therapeutic target in cancer, as tumors with BRCA mutations would be susceptible to loss of RAD52, while normal tissues with an intact HR pathway should survive without it. This genetic relationship is known as synthetic lethality, where loss of function of each gene on its own still results in viable cells, while loss of both results in cell killing. Synthetic lethality is an attractive strategy for targeted therapy, as it capitalizes on natural genetic relationships for tumor-specific kill instead of damaging agents, which can cause toxicity to both healthy cells and tumor cells
[21][50].
In the context of DNA repair, synthetic lethality can especially be exploited when tumor cells deficient in one repair pathway are “addicted” to a backup pathway. Loss of the backup pathway then leads to toxic intermediate structures and cell death. The most common example of pharmacological exploitation of synthetic lethality is poly-ADP-ribose polymerase (PARP) inhibition in HR-deficient cells. Since PARP can recognize single-stranded breaks (SSBs), inhibiting its function would leave unrepaired SSBs which could be converted to DSBs during replication. In HR-defective cells, these breaks would either be left unrepaired or would be channeled to error-prone repair processes. The first FDA-approved PARP inhibitor, olaparib, was authorized for treatment in a subset of BRCA-mutated cancers in 2014, followed by the development and approval of a new generation of inhibitors
[50]. While these inhibitors have been effective in some contexts, they have also been shown to cause global toxicity, likely because of the role of PARP proteins in cellular processes other than DNA repair
[51]. The effectiveness of PARP inhibitors supported the use of synthetic lethality as an indicator for therapeutic success, but their adverse effects have inspired some to look to other synthetically lethal interactions for the treatment of HR-deficient tumors.
As homozygous loss of RAD52 caused no effect in mice, it is unlikely to cause toxicity to healthy tissues in humans. Its effect in HR-deficient tumors, on the other hand, is significant and well-characterized. While initially described as being synthetically lethal with BRCA2, it has since been found to also lead to cell death in the absence of BRCA1, PALB2, and RAD51 paralogs
[20][21][52]. Additionally, recent super resolution microscopy data that mapped the spatiotemporal kinetics of HR repair showed that RAD52 and BRCA2 have distinct functions in the RAD51 nucleofilament formation and subsequent homology search. While in the absence of RAD52, BRCA2 was able to compensate for RAD52, the converse was not true
[53]. This may explain why RAD52 loss in a BRCA2-proficient context has no observable effect. In the context of BRCA1 loss, one proposed mechanism for synthetic lethality with RAD52 was reliant on the endonuclease EEPD1, which recognizes stalled forks and cleaves them into a DSB. In the absence of BRCA1 and RAD52, accumulation of unrepaired breaks at collapsed forks, which are normally repaired by HR, results in cell death. The synthetic lethality is rescued by EEPD1 co-depletion, as the forks are restarted or repaired by other mechanisms which, while error-prone, do not involve these toxic intermediates
[54].
Because RAD52 loss is so detrimental to BRCA-deficient cells, many groups have conducted screens to identify compounds that could serve as inhibitors of RAD52 function. The inhibitors target RAD52 in one of two ways: either by inhibiting its annealing activity by competitively binding to the ssDNA-binding domain, or by interfering with its ability to multimerize and form the ring structure required for its activity
[50][55][56][57][58][59]. Some of these compounds may be useful in HR-deficient cancers, either as monotherapy or in combination with other agents. One such inhibitor, 6-OH-dopa, was shown to have an additive effect on BRCA-deficient cancer cells treated with PARP inhibitors, which they termed “dual” synthetic lethality
[60]. Addition of RAD52 inhibitors could decrease the necessary dosage of PARP inhibitors, hopefully reducing their toxicity.
RAD52 inhibitors may also be useful outside the realm of BRCA deficiency, as depletion of RAD52 was shown to decrease tumor volume in FANCM-deficient tumors in mice. This interplay of RAD52 and FANCM was important in the repair and stability of common fragile sites, and relied on the translocase activity of FANCM rather than its association with the rest of the Fanconi anemia (FA) core complex
[61]. It remains to be determined whether any of the other myriad roles of RAD52 represent a new context in which an inhibitor may be useful.