1000/1000
Hot
Most Recent

The coronavirus disease 2019 (COVID-19) outbreak, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global ongoing pandemic. Timely, accurate and non-invasive SARS-CoV-2 detection in both symptomatic and asymptomatic patients, as well as determination of their immune status, will facilitate effective large-scale pandemic control measures to prevent the spread of COVID-19. Saliva is a biofluid whose anatomical source and location is of particularly strategic relevance to COVID-19 transmission and monitoring. This review focuses on the role of saliva as both a foe (a common mode of viral transmission via salivary droplets and potentially aerosols) and a friend (as a non-invasive diagnostic tool for viral detection and immune status surveillance) in combating COVID-19.
The timing (highest viral titres) and specimen collection sources can significantly influence the diagnostic sensitivity of SARS-CoV-2 detection tests. One study reported that oropharyngeal swabs (n = 398) were more often used than nasopharyngeal swabs (n = 8) in China during the COVID19 outbreak; however, SARS-CoV-2 RNA was detected in only 32% of oropharyngeal swabs [1]. On 19 March 2020, the World Health Organisation (WHO) recommended that both upper (nasopharyngeal and oropharyngeal swabs) and lower (sputum, bronchoalveolar, or lavage endotracheal aspirate) respiratory specimens should be collected; however, upper respiratory samples may fail to detect early viral infection and the collection of lower respiratory specimens increases biosafety risk to healthcare workers via aerosol/droplets formation. As the SARS-CoV-2 virus shedding progresses, additional samples sources, such as stool, saliva, and blood, can be used as alternatives, or combined with respiratory specimens. However, only 15% of patients hospitalised with pneumonia had detectable SARS-CoV-2 RNA in serum [2], and 55% of patients showed positive SARS-CoV-2 RNA in fecal samples [3]. Conversely, in saliva samples, it was reported from different clinical studies that 87%, 91.6%, and 100% of COVID-19 patients were identified as being viral positive, respectively [4][5][6], suggesting that saliva is a powerful specimen source for the diagnosis of the SARS-CoV-2 virus.
Saliva also represents an attractive biofluid source option for the detection of SARS-CoV-2, due to being non-invasive, easy-to-access, and low-cost, as well as having the ability to “mirror” systemic and local disease status [7]. It is well-known that saliva harbors a wide range of circulatory components (Figure 1), such as pro-inflammatory cytokines [8][9], chemokines [10], matrix metalloproteinases [11][12], mitochondrial DNA [13], genomic DNA [14], bacteria [15], SARS-CoV and SARS-CoV-2 virus [16][4][5], SARS-CoV antibodies [16], miRNAs [17], and extracellular vesicles (EVs) [18]. Furthermore, saliva samples can be stored at –80 °C for several years with little degradation [19]. It is preferable to aliquot and freeze the samples to avoid freeze–thaw cycles. For salivary RNA research, it was discovered that saliva samples can be stored in Trizol for more than two years at –80 °C without adding RNase inhibitors [20][21], suggesting such specimens can be used for future diagnostics. Thus, saliva may be a valuable specimen to collect in COVID-19 patients at different time points during disease onset progression and follow-up. Indeed, saliva may be useful for both diagnosing the presence and sequelae of COVID-19 infection, as well as identifying and tracking the development of immunity to the virus.
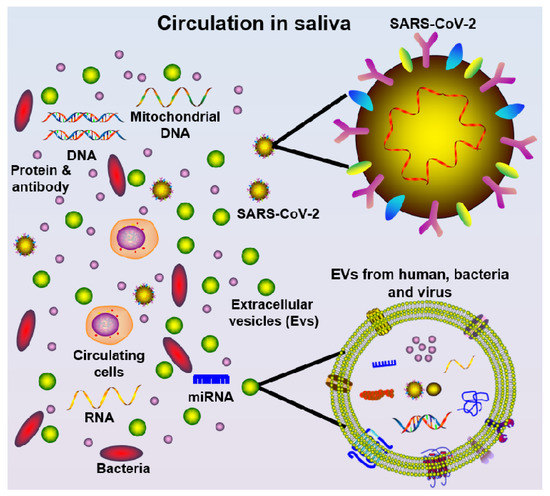
Figure 1. Schematic diagram of saliva components, including cells, mitochondrial DNA, DNA, protein/antibody, bacteria, miRNA, extracellular vesicles (EVs, from multiple oral cavity resident species), and SARS-CoV-2 virus.
Saliva has been widely investigated as a potential diagnostic tool for chronic systemic and local (oral) diseases [7], with less attention given to its utility in acute infectious diseases, such as COVID-19. The salivary gland can be infected by SARS-CoV-2 virus resulting in the subsequent release of viral particles or antibodies into saliva, as evidenced in Rhesus macaque primates where salivary gland epithelial cells were the first target cells for SARS-CoV infection [16]. This is likely to be facilitated by the high expression of hACE2 (SARS-CoV-2 receptor) on the epithelial cells of the oral mucosa, as demonstrated using single-cell RNA sequencing [22].
Saliva and throat wash (by gargling 10 mL saline) samples from 17 SARS-CoV patients were found to be SARS-CoV RNA positive, with the highest detection rate a median of four days after disease onset and during lung lesion development [23]. Saliva samples from 75 patients successfully validated saliva as a viable biosample source for COVID-19 detection when compared to nasopharyngeal or oropharyngeal swabs [24].
At present, only three clinical studies (Table 1) and one animal model have investigated the use of salivary diagnostics for COVID-19. SARS-CoV-2 was detected in self-collected saliva (by asking the patients to expectorate saliva) in 11 out of 12 confirmed cases [5]. Another recent study found that 100% of COVID-19 patients (n = 25) were detected as viral positive in drooling saliva samples [6]. Further, in a cohort of COVID-19 positive patients, it has been demonstrated that 87% of posterior oropharyngeal (deep throat) saliva samples were detected viral positive (n = 23), and serial respiratory viral load of SARS-CoV-2 was detected from week 1 and up to 25 days after symptom onset, while serum (n = 16) samples showed positive RT-qPCR detection only 14 days after symptom onset [4]. Additionally, Kim et al. demonstrated that SARS-CoV-2-infected ferret animals shed virus in nasal washes, saliva, urine, and feces up to eight days post-infection and ferret-to-ferret transmission occurred only two days post-contact [25]. Notwithstanding the limitations of small sample size and lack of detailed saliva collection methodology, these studies nevertheless imply that saliva is a promising non-invasive alternative specimen for SARS-CoV-2 diagnosis. Further investigations are required to explore the potential role of saliva for COVID-19 detection in both symptomatic and asymptomatic patients.
Table 1. Current clinical research finding using salivary diagnosis for COVID-19.
| Sample Size Age (Years) |
Sample Source | Diagnosis Technique | Diagnosis Efficiency in Saliva | Reference | |
|---|---|---|---|---|---|
| To et al. | 10 Female, 13 Male Median: 62 (37–75) |
Posterior oropharyngeal saliva | RT-qPCR | 87% of patients were viral positive | [4] |
| To et al. | 5 Female, 7 Male Median: 62.5 (35–75) |
Saliva from throat | RT-qPCR | 91.7% of patients were positive | [5] |
| Azzi et al. | 8 Female, 17 Male Mean ± standard deviation: 61.5 ± 11.2 |
Drooling saliva | RT-qPCR | 100% of patients were viral positive | [6] |
In summary, the current gold standard diagnostic test is RT-qPCR to detect SARS-CoV-2 RNA which takes approximately 48 h to obtain the test results. More new tests with higher sensitivity and specificity need to be appropriately validated before being implemented into the current routine diagnosis.
From early reports on the clinical characteristics of COVID-19, it is now apparent that not all people exposed to SARS-CoV-2 are infected and not all infected patients develop severe symptoms [26]. Indeed, three broad presentations of SARS-CoV-2 infection can be characterised: (i) an asymptomatic incubation stage with or without detectable virus; (ii) non-severe symptomatic presentation with confirmed presence of virus; and (iii) a severe respiratory symptomatic stage with high viral load [27]. Determining the immune status of an individual is likely to become increasingly critical as the COVID-19 pandemic progresses, because from a prevention perspective, individuals at stage I (the stealth carriers or the super spreaders), are particularly important because they may spread the virus unknowingly.
Two stages of the immune response during COVID-19 disease progression have been proposed [28]: (1) Immune-defense-based protective phase: elimination of SARS-CoV-2 virus by an individual’s adaptive immune response; and (2) inflammation-driven phase: when the protective immune response is impaired and prolonged propagated virus load leads to an adverse inflammatory response in organs with high hACEs expression. Indeed, a likely pathogenic mechanism of SARS-CoV-2 is overactivation of T cells with an increase in CD4+ T Helper cells and enhanced cytotoxicity of CD4+ and CD8+ T cells [29], which leads to an imbalanced pro-inflammatory and anti-inflammatory cytokine response and severe immune injury in susceptible patients [30]. Although this concept needs to be confirmed by more clinical research, it may provide useful research directions to tackle COVID-19.
During the previous SARS outbreak, a common transmission pattern hypothesis was that SARS-CoV virus silently infected asymptomatic patients, which may have led to population immunity against infection (herd immunity) that may explain the eradication of the virus, although this is yet to be confirmed [31]. Although a study suggests that coronavirus antibodies are highly prevalent in the general population after exposure to four non-SARS coronavirus strains [32], there is no definitive evidence on whether permanent immunity would be generated against other CoV species, such as SARS-COV-2. Notably, after SARS-CoV infection in a murine model, the production of SARS-CoV-specific serum IgG and secretory immunoglobulin A (sIgA) were detected in saliva following intranasal immunisation [33].
In relation to COVID-19, intensive care unit (ICU) patients had higher plasma levels of pro-inflammatory cytokines, including IL-2, IL-7, IL-10, GSCF, IP10, MCP-1, MIP-1A, and TNF-α, compared with non-ICU patients [2], suggesting the emergence of a robust immune-inflammatory response in severe symptomatic COVID-19 patients. Importantly, several studies have demonstrated that COVID-19 patients developed IgG and IgM antibodies against SARS-CoV-2 in blood samples. Both IgG and IgM antibodies against the SARS-CoV-2 nucleoprotein and spike receptor-binding domain were increased in serum at day 10 after symptom onset for up to three weeks [4]. A point-of-care lateral flow immunoassay (LFIA) test product (VivaDiag COVID-19 IgM/IgG Rapid Test) was designed to detect IgM and IgG in blood samples of COVID-19 patients in 15 min [34]. However, the sensitivity of the VivaDiag COVID-19 IgM/IgG Rapid Test was only 18.4% in blood samples of acute COVID-19 patients from the emergency department [35], suggesting that serological tests require more research before being deemed suitable for routine diagnosis. Additionally, the seroconversion rate for total antibodies, IgM, and IgG were shown to be 93.1%, 82.7%, and 64.7%, respectively, in hospitalised COVID-19 patients, peaking 7–14 days after symptom onset [36]. Given the non-invasive and cost-effective nature of saliva collection, it would be important to investigate whether this immunity detection is feasible in saliva samples as a tool for facilitating the testing of COVID-19 immunity at the population-level.