The upregulated expression of both NUB1 and FAT10 has been observed in various cancers. NUB1 protein binds to FAT10 non-covalently to promote FAT10 degradation. An overexpressed FAT10 stimulates nuclear factor-kappa β, activates the inflammatory pathways, and induces the proliferation of cancer. The FAT10 protein interacts with the mitotic arrest deficient 2 protein, causing chromosomal instability and breast tumourigenesis. FAT10 binds to the proliferating cell nuclear antigen protein and inhibits the DNA damage repair response. In addition, FAT10 involves epithelial–mesenchymal transition, invasion, apoptosis, and multiplication in hepatocellular carcinoma.
1. Introduction
Identifying biomarkers could aid in clinical decision making since it allows personalised medicine in oncology. There are two major types of biomarkers: predictive markers, which direct the use of modified therapies, and prognostic markers, which assist the evaluation of malignancy features and clinical trial planning. With the availability of biomarkers, managing cancer therapy has revolved around the tenets of genetics and proteomics. Since then, there is a growing interest in expanding the application of biomarkers in managing cancer patients. Additionally, the clinical potential of monitoring disease using immunohistochemistry is well established, particularly in the analysis of receptor status in breast cancer. However, findings on the use of NEDD8 ultimate buster 1 (NUB1) and F-adjacent transcript 10 (FAT10) proteins as a prognostic biomarker remain scarce. To date, oncologists still find it hard to anticipate the chemotherapy response in many cancer patients in selecting the most efficacious treatment for tumours based on their biology. This literature review is based on the search of the electronic database PubMed.
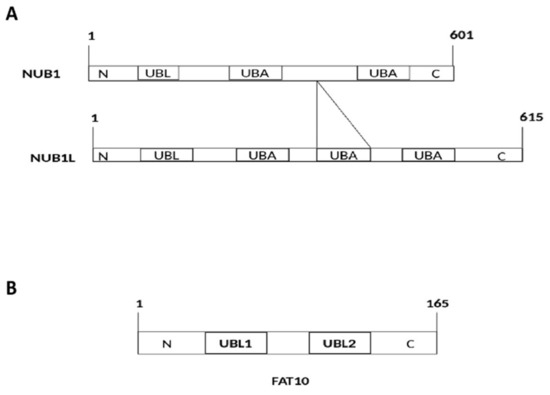
The NUB1 protein, discovered by Yeh et al.
[1], consists of 601 amino acid residues (
Figure 1A) with a molecular weight of 69.1 kDa. Its splice variant, the NEDD8 ultimate buster-1 long (NUB1L) protein, interacts with FAT10 protein through non-covalent bonds. Both NUB1 and NUB1L are interferon-inducible proteins that trigger the degradation of neddylation proteins by the ubiquitin–proteasome system
[1]. They bind to the proteasome regulatory particle base subunit ribophorin 10 (RPN10) of the 26S proteasome to facilitate the degradation of ubiquitin-like (UBL) proteins, i.e., FAT10
[1].
Figure 1. (A) Structural and functional domains of NEDD8 ultimate buster-1 (NUB1) and NEDD8 ultimate buster-1 long (NUB1L). UBL: Ubiquitin-like domain (interacts with proteasome); UBA: Ubiquitin associated domain (interacts with ubiquitin-related enzymes). An additional UBA (14 amino acids) domain in NUB1L. (B) Structural and functional domains of F-adjacent transcript 10 (FAT10). FAT10 comprises 165 amino acids. UBL: Ubiquitin-like domain (interacts with VWA domain of 26S proteasome).
2. Interaction of FAT10 and NUB1
Meanwhile, the subunit RPN10 (S5a) of the 26S proteasome serves as an anchoring site for NUB1L, FAT10, and polyubiquitin
[2]. Specifically, FAT10 binds to the von Willebrand A (VWA) domain of RPN10 via its
C-terminal UBL domain and to the UBA domains of NUB1L through its
N-terminal UBL domain. Consequently, NUB1L interacts with the VWA domain of Rpn10 protein by its
N-terminal UBL domain. Thus, FAT10 can bind to RPN10 either directly or via NUB1L
[2][3].
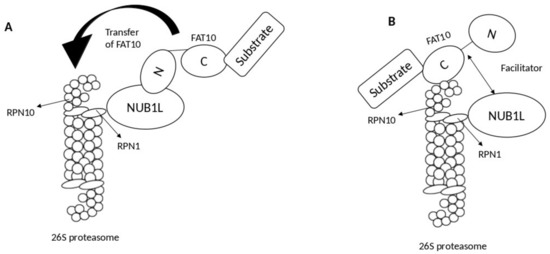
Two models describe the role of NUB1L as a soluble receptor: the transfer model and the facilitator model. In the transfer model, NUB1L interacts with the
N-terminal domain of FAT10 and subsequently transfers it to RPN10 upon interacting with ribophorin 1 (RPN1)
[3]. By contrast, in the facilitator model, NUB1L, with the 19S regulator of the proteasomal subunit RPN1 (S2), triggers conformational modifications in both RPN1 and RPN10 for interacting with FAT10 and its subsequent conjugation with RPN10 (
Figure 2)
[3].
Figure 2. Proposed models show the mechanism of NEDD8 ultimate buster-1 long (NUB1L) in accelerating F-adjacent Transcript 10. (FAT10) degradation by the proteasomal system. (A) Model A: the transfer model; NUB1L acts as a receptor and binds to N-terminal of FAT10 and transfer it to ribophorin 10 (RPN10) upon binding to ribophorin 1 (RPN1). (B) Model B: the facilitator model; NUB1L acts as a facilitator by binding with RPN1 and later inducing conformational changes within both RPN10 and RPN1 so FAT10 could bind to RPN10, and its degradation can occur.
3. NUB1 Protein Actions in Cancer
NUB1 has been examined in various cancer cell lines, including renal cell carcinoma (RCC), cervical adenocarcinoma, neuroblastoma, rectal adenocarcinoma, and malignant lymphoma
[4]. Overexpression of NUB1 in cancer cells is associated with IFN-α induced antimitogenic activities
[5]. NUB1 displays anticancer potential in RCC cell lines with S-phase transition and apoptotic properties by cyclin E and p27; p27 inhibits the cyclin E-CDK2 complexes and hence hinders the progression from the G
1 phase to the S-phase of the cell cycle. Thus, NUB1 protein causes apoptosis and cell-cycle arrest in RCC cells. Overexpression of NUB1 also prevents the proliferation of interferon-α (IFN-α)-resistant RCC cells in vitro
[5].
Meanwhile, tumour metastasis and invasion are generally characterised by epithelial–mesenchymal transition (EMT). NUB1 protein decreases the expression of EMT proteins, i.e., N-cadherin, matrix metalloproteinase-2 (MMP2), and vimentin, while significantly overexpressing E-cadherin, which is an epithelial marker
[6]. This mechanism was identified, and it showed that NUB1 proteins prevent malignancy in gastric cancer cell lines in vitro
[6].
Another group of proteins related to the malignancy of cancer is the S-phase kinase-associated protein 2 (SKP2) that belongs to the protein complex of Skp, Cullin, and F-box (SCF complex). Deregulating SKP2 or SCF family proteins degrades the p27
Kip1 in gastric cancer to trigger the malignancy
[6][7]. Since NUB1 controls the cell cycle progression via the upregulation of p27
Kip1 [6], overexpression of NUB1 and NUB1L may prevent the activities of SCF family proteins via suppressing the ligase activity of SCF complexes to treat malignant phenotypes in cancer cells
[8].
Additionally, the neural precursor cell expressed developmentally downregulates protein 8 (NEDD8), a UBL protein that controls many essential biological functions in the regulation of cell cycles
[9]. Although the neddylation of proteins in human cells is inevitable, overexpression of NEDD8 may trigger abnormal effects in cells, leading to the development of cancer
[10]. To date, NUB1 and NUB1L have been shown to interact with NEDD8 protein to initiate its degradation through the proteasomal system
[10][11]. Specifically, NUB1/NUB1L non-covalently binds to NEDD8 through their UBA domains to prevent its aberrant effects in cells and, ultimately, the development of cancer
[10].
4. FAT10 Protein Actions in Cancer
As a UBL, FAT10 protein regulates cell survival and cell growth. The mRNA and protein of FAT10 are overexpressed in tumours
[12]. FAT10 also governs the cellular immune system, and it occurs abundantly in tissues, such as the spleen, lymph nodes, and thymus in humans
[13]. The expression of FAT10 is upregulated through the actions of proinflammatory cytokines, i.e., tumour necrosis factor-a (TNF-a) and interferon g (IFNg). An in vivo study showed that IFNγ and TNF-a were responsible for upregulating the mRNA expression of FAT10 in hepatocytes of alcoholic cancer patients
[14]. Meanwhile, the degradation of FAT10 could occur only through the 26S proteasome
[14]. The upregulated expression of FAT10 accelerates the proliferation and progression of hepatocellular carcinoma, bladder cancer, colon cancer, and cervical cancer (
Table 1)
[15][16].
Table 1. The actions of FAT10 protein and its involvement with target proteins and pathway in various cancer types; Hepatocellular carcinoma (HCC); 78-kDa glucose-regulated protein (GRP78); Nuclear factor-κB (NF-κB); zinc finger E-box binding protein 2 (ZEB2); mitotic arrest deficient 2 (MAD2); Proliferating cell nuclear antigen (PCNA); SMAD Family Member 2 (SMAD2); Non-small cell lung cancer (NSCLC).
|
Cancer Type
|
Remarks
|
References
|
|
Hepatocellular carcinoma (HCC)
|
GRP78 protein increases FAT10 protein expression via direct activation on the NF-κB pathway.
|
[17]
|
|
Breast cancer
|
FAT10 protein induces pro-metastasis effect with the help of ZEB2 overexpression.
|
[18]
|
|
Bladder cancer
|
FAT10 protein non-covalently binds to Survivin protein to inhibit ubiquitin-mediated degradation.
|
[19]
|
|
B-cell non-Hodgkin lymphomas
|
FAT10 protein non-covalently binds to MAD2 protein to maintain mitosis.
|
[20]
|
|
Colorectal cancer, HCC, Gastric cancer
|
FAT10 protein disrupts the DNA damage repair response via modification of PCNA protein.
|
[21][22][23]
|
|
NSCLC
|
FAT10 causes NSCLS malignancy via interaction with NF-κB signalling pathway.
|
[24]
|
|
Glioma
|
FAT10 protein increases phosphorylation of SMAD2 protein, which triggers FAT10 induced oncogenic activities.
|
[25]
|
|
Neuroblastoma
|
FAT10 protein stabilises the survivin protein via non-covalent binding.
|
[26][27]
|
It is generally thought that the glucose-regulated protein 78 (GRP78) could phosphorylate and stimulate the P13K/AKT signalling pathway to promote cancer growth
[17][28]. The GRP78 protein increases the FAT10 protein expression via the NF-κB pathway, while the FAT10 gene reduces the activity of the tumour-suppressor gene, p53. Thus, the GRP70-NF-κB-FAT10 axis could be a therapeutic target to treat hepatocellular carcinoma (HCC), and further proliferation of HCC cells could be inhibited or reduced
[20]. FAT10 protein activates the NF-κB signalling pathway to boost the proliferation of cancer cells
[29][30], and in turn, the NF-κB signalling pathway upregulates the pathogenicity and proliferation of HCC
[31][32].
Furthermore, FAT10 protein is upregulated in the hepatitis B virus (HBV) in association with the HCC tissues
[13][33]. The protein stimulates the signalling pathway of protein kinase B/glycogen-synthase kinase 3 Beta (PKB/GSK3β) linked to invasion, EMT, apoptosis, and proliferation in HCC. This finding further shows that FAT10 could serve as a potential biomarker and prospective target to diagnose and treat HBV-related HCC
[34].
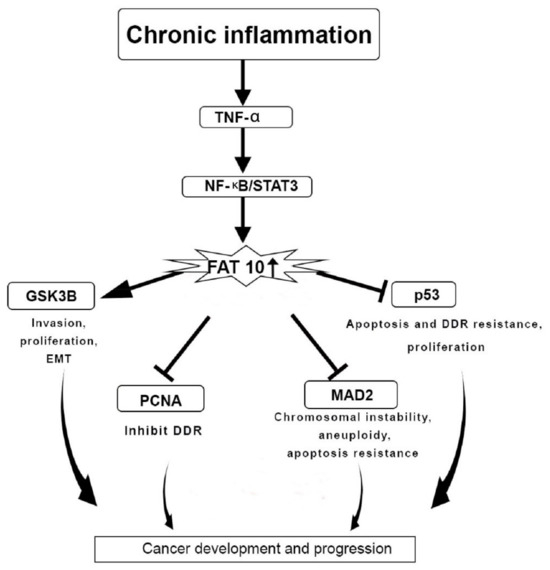
One of the primary causes of cancer progression is chronic inflammation
[35]. TNF-α contributes to cancer development by generating chronic inflammation
[36]. TNF-α triggers the expression of the FAT10 gene via the TNF receptor 1, inducing the inflammatory signalling pathway of NF-κB in cancer cell lines. TNF-α is also responsible for disrupting the mitotic phase during the cell cycle. This phenomenon could be prevented through the actions of FAT10 protein that induces the activity of TNF-α during cancer pathogenesis by disrupting the cell cycle, chromosomal instability, and inhibition of apoptosis
[29].
The gene expression of FAT10 is linked to the expression of the signal transducer and the activation of the gene transcription 3 (STAT3). The expression of FAT10 is synergistically triggered through the activation of NF-κB (
Figure 3)
[30]. The STAT3 gene stabilises NF-κB on the promoter region of FAT10 to enhance the expression FAT10 gene
[30][37]. The p53 protein is responsible for the degradation of FAT10 protein. Therefore, the interaction of FAT10-p53 is essential to halt cancer progression
[38]. There is a transcriptional regulation between STAT3 and NF-κB that impacts FAT10 expression to inhibit p53 expression and hence support cancer and inflammation progression (
Figure 3)
[29].
Figure 3. FAT10 protein actions in cancer development and progression. Chronic inflammation regulates the F adjacent transcript 10 (FAT10) gene expression as well as downstream targets through tissue necrosis factor-α (TNF-α) and Nuclear factor- κB/Signal transducer and activator of transcription 3 (NF-κB/STAT3) pathway, which is responsible for upregulating the FAT10 gene expression. Overexpression of FAT10 further mediates the epithelial–mesenchymal transition (EMT), invasion, proliferation, chromosomal instability, DNA damage repair (DDR) resistance, and apoptosis. This leads to cancer development and progression; Glycogen synthase kinase 3 beta (GSK3β); Proliferating cell nuclear antigen (PCNA); mitotic arrest deficient 2 (MAD2); Tumor protein p53 (p53).
5. Use of NUB1 and FAT10 as Biomarkers in a Clinical Setting
Several studies investigated the correlation between the NUB1 and FAT10 proteins and the survival probability among cancer patients.
Table 2 summarises the findings that identified NUB1 and FAT10 as prognostic and predictive biomarkers. Although the concentration of NUB1 mRNA is higher in cancer cells, the depletion of NUB1 protein could lead to G
0/G
1 cell cycle arrest in vitro. The knockdown of NUB1 prevents the growth of MDA-MB-231 cell lines in vitro. The cell cycle arrests lead to the death of the breast cancer cells because of the accumulation of p21/p27 proteins in NUB1-depleted cells
[39].
Table 2. Summary of translational NEDD8 ultimate buster 1 (NUB1) and F-adjacent transcript 10 (FAT10) studies that examine the correlation of protein expression to the survival probability of cancer patients. NUB1 and FAT10 protein statuses are identified as prognostic and potentially predictive biomarkers; NEDD8 ultimate buster 1 long (NUB1L); Non-small-cell lung cancer (NSCLC).
|
Type
|
Human Sample Types
|
Sample Size
|
Antibody Clone and Host Species
|
Method of Detection and Biomarker Type
|
Findings
|
References
|
|
Anti-NUB1 and -NUB1L
|
Gastric cancer patients
|
116
|
Ab38438
(Rabbit polyclonal)
|
Immunohistochemistry/
Prognostic
|
Reduced NUB1 level associated to poor prognosis of gastric cancer.
p < 0.05; HR: 0.33 (0.20–0.54)
|
[6]
|
|
Breast cancer patients
|
114
|
4H2 (Mouse Monoclonal Antibody)
|
Immunohistochemistry/
Prognostic
|
Low cytoplasmic
NUB1 protein level exerts poorer overall survival.
p = 0.048, HR: 1.779 (1.006–3.346)
|
[39]
|
|
Anti-FAT10 antibody
|
Bladder cancer samples
|
133
|
MBS4750652
(Rabbit Polyclonal Antibody)
|
Immunohistochemistry/
Prognostic
|
Higher FAT10 expression in bladder cancer tissues had poorer survival than those with lower FAT10 expression.
p = 0.002; HR:?
|
[19]
|
|
Non-small cell lung carcinoma (NSCLC) samples
|
45
|
sc-133199
(mouse monoclonal antibody)
|
Immunohistochemistry/
Prognostic
|
High FAT10 expression confers quick chemoresistance than the lower FAT10 expression group.
p = 0.001; HR:?
|
[24]
|
|
Breast cancer tissues
|
120
|
ab168680 (Mouse polyclonal antibody)
|
Immunohistochemistry/
Prognostic
|
FAT10 overexpression leads to poor prognostic factor for poorer outcomes of patients with breast cancer.
p < 0.05; HR:1.563 (1.232–2.531)
|
[18]
|