Osteosarcoma (OS) is a highly aggressive malignant bone tumor, frequently occurring in children and adolescents with an annual incidence of over three per million worldwide.
1. Introduction
Osteosarcoma (OS) is a highly aggressive malignant bone tumor, frequently occurring in children and adolescents with an annual incidence of over three per million worldwide
[1][2][3]. OS represents different pathological entities based on clinical, radiological, and histopathological features. For instance, based on histopathological features, osteosarcoma can be classified into distinct subtypes with the osteoblastic, chondroblastic, and fibroblastic OS, respectively, being the most common
[4].
Nowadays, various clinical practices for OS have been notably implemented, including chemotherapy, radiotherapy, surgery, and targeted therapy; yet, the prognosis for OS still remains poor
[5][6]. In fact, approximately 20% of patients showed clinical metastasis at presentation, with a 5-year survival rate less than 30%
[7]. For this reason, OS strongly demands reliable, non-invasive, and clinically useful biomarkers.
In contrast to conventional biopsy, the liquid biopsy of tumor components in blood represents a simple and rapid test, easily performed, and requiring a small amount of sample (usually 10–15 mL of blood). Presently, however, the usefulness of alkaline phosphatase (ALP) and lactate dehydrogenase (LDH) as laboratory markers for OS is still considered controversial
[8][9]. Likewise, studies have shown that programmed cell death 1 ligand-1 (PD-L1) and bone resorption markers, such as b-isomerized C-terminal telopeptides (b-CTx) and total procollagen type 1 amino-terminal propetide (tP1NP), still require more investigation before being able to conclude their potential value as biomarkers for OS
[6][10][11][12].
Recently, circulating biomarkers, such as circulating tumor cells (CTCs) and different forms of circulating-free and extracellular vesicle/platelet-encapsulated non-coding RNA, including microRNA (miRNA) and long non-coding RNA (lncRNA), have emerged as novel promising diagnostic, prognostic, or predictive biomarkers in the clinical management of patients with OS
[13][14][15][16][17][18].
Although CTCs may provide tumor-specific genomic, transcriptomic, and proteomic information, their analysis requires a large volume of fresh blood and it is laborious and expensive. On the other hands, the use of circulating ncRNAs, in spite of some obvious limitations, is more accessible, cheaper, and has shown potential as a precision medicine biomarker
[19]. Early studies on circulating RNAs focused on the relevance of miRNAs. However, the current search for novel OS biomarkers has possibly shifted to lncRNAs due to their relative abundance and higher stability with respect to miRNAs
[14].
Interestingly, a number of circulating lncRNAs, whose expression in liquid biopsy correlate with that of cancer tissues, have emerged as novel diagnostic or prognostic markers for several types of cancer
[20][21][22][23]. However, the role of circulating lncRNAs as biomarker for OS is still elusive.
2. Long Non-Coding RNA Structures and Functions
LncRNAs are conventionally classified as transcripts longer than 200 nt with no or low coding potential
[24][25][26]. Similar to protein-coding transcripts, the transcription of lncRNAs is dependent on histone-modification-mediated regulation, and lncRNA’s transcripts are processed by the canonical spliceosome machinery. Overall, lncRNA genes show fewer exons than mRNAs, and appear to be under a weaker selective pressure during evolution. Moreover, some lncRNAs are expressed at levels lower than those of mRNAs and in a more tissue- and cell-specific manner, while others are known to be fairly abundant and are expressed in diverse cell types, such as the “house-keeping” genes
[27].
Of tens of thousands of metazoan lncRNAs discovered from cDNA libraries and RNAseq data by high throughput transcriptome projects, only a handful of lncRNAs have been functionally characterized. The investigations on this small cohort of lncRNAs have demonstrated that these noncoding transcripts can serve as scaffolds or guides to regulate protein–protein or protein–DNA interactions
[28][29][30][31] or can modulate post-translational modification of nonhistone proteins
[32]. Moreover, lncRNAs are capable of controlling microRNAs (miRNAs)
[33][34][35], and function as enhancers to influence gene transcription, when transcribed from the enhancer regions (enhancer RNA)
[36][37][38] or their neighboring loci (noncoding RNA activator)
[39][40].
Several lines of evidence have shown that lncRNAs are capable of influencing different cellular functions that are critical to tumorigenesis, such as cell proliferation, differentiation, migration, immune response, and apoptosis
[41][42][43][44][45][46][47]. Furthermore, lncRNAs have been found to act as tumor suppressors or oncogenes
[48][49][50][51], and, of note, a number of lncRNAs have been reported to be significantly deregulated in tumors
[52][53][54][55].
3. Origin of Circulating lncRNAs
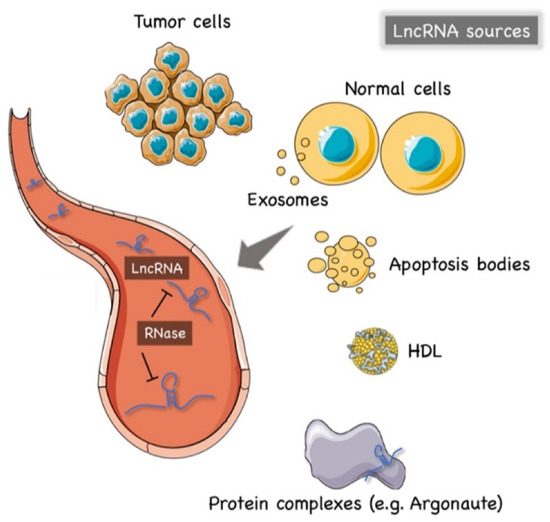
The precise mechanism of lncRNAs release into the extracellular environment is not completely understood. Hypotheses have arisen that tumor cells, cancer-adjacent normal cells, immune cells, and other blood cells may all release lncRNAs
[56][57], as shown in
Figure 1. A few studies reported that lncRNAs can be encapsulated into membrane vesicles, such as exosomes or microvesicles (EV), prior to being secreted extracellularly. In such a conformation, the circulating lncRNAs have shown a higher degree stability, probably due to EVs offering protection against the nuclease-mediated degradation that may occur in the extracellular space and in body fluids
[20][58][59] (
Figure 1). On the other hand, other studies have suggested that the secretion pathway of lncRNAs may also occur in a similar manner to that for miRNAs. As such, lncRNAs might also be released into body fluids in an EV-independent fashion as complexes with high-density lipoproteins (HDLs) or protein Argonaute 2 (AGO2)
[60] (
Figure 1).
Figure 1. The origin of circulating lncRNAs. Two major sources of circulating lncRNAs have been postulated so far. LncRNAs can be encapsulated in extracellular vesicles (EV), predominantly exosomes. On the other hand, lncRNAs can also be released from live cells in an EV-independent fashion, thus, similar to circulating miRNAs, circulating lncRNAs might be detected in complexes with protein or high-density lipoproteins (HDL). The latter mechanism is likely to offer = circulating lncRNAs less protection against ribonucleases that are normally present in the extracellular space and body fluids. Created by Servier Medical Art (http://smart.servier.com/ (accessed on 15 May 2021)), licensed under Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/ (accessed on 15 May 2021)).
The hypothesis of an EV-independent mechanism for lncRNAs secretion might seem less likely given the high abundance of ribonucleases in serum, plasma, and other bodily fluids that can dramatically affect the stability of lncRNAs in the extracellular environment. However, one can speculate that circulating lncRNAs can be capable of resisting the RNase activity through modifications such as methylation, adenylation, and uridylation
[61] or via the formation of higher order structures
[62].
4. Detection Methods of Circulating lncRNAs
Difference sources of liquid biopsy (i.e., whole blood, plasma, serum, urine, and gastric juice) can be used to quantify circulating lncRNAs. However, due to the possibility of blood cell RNA contamination, whole blood is the less recommended option so far
[63]. In addition, EDTA-anticoagulant collecting tubes have been suggested to be more suitable for the analysis of circulating lncRNAs
[57]. Of note, some studies have found that lncRNAs remained stable in plasma even under multiple cycles of freeze–thaw, incubation at 45 °C, or storage at room temperature for as long as 24 h
[56].
Overall, the methods to extract circulating lncRNAs can be divided into two major groups: guanidine/phenol/chloroform-based and column-based protocols. The column-based method is currently considered more reliable, since organic and phenolic contaminants in TRIzol-based methods might invalidate results
[64].
Regarding the measuring and normalization methods, some studies have suggested that the use of an equal volume of input RNA sample may be more accurate than an equal amounts of RNA measured using a NanoDrop spectrophotometer since many diseases, including cancer, may indeed release an higher degree of RNAs into body fluids than healthy control groups, leading to a significantly higher level of circulating RNA in cancer patients that causes misleading results
[64].
To date, quantitative real-time PCR (qRT-PCR) is still considered the gold standard for quantitative expression analysis of lncRNAs, including circulating lncRNAs
[65]. Microarrays and whole transcriptome analysis (RNA-seq) still have limited uses in this field. In fact, the high throughput potential of microarrays relies on a reference database of targets, which in the case of circulating lncRNAs, is still very limited
[21]; the RNA-seq requires huge amounts of starting RNA samples. Additionally, RNA-seq is currently expensive and needs special equipment and/or expert bioinformaticians
[64], whereas, the targeted-approach of qRT-PCR is still more accessible, and saves money and time. Accordingly, qRT-PCR can be divided into relative and absolute analyses. In relative quantification methods, the choice of endogenous controls is critical to properly normalize the expression levels. In this regard, it must be noted that no systematic evaluation of reference genes for serum lncRNA has yet been reported, posing some limitations for the relative qRT-PCR method in the analysis of lncRNAs from a liquid biopsy.