Circular RNAs (circRNAs) are a new class of endogenous non-coding RNAs with covalent closed loop structure. Researchers have revealed that circRNAs play an important role in human diseases. As experimental identification of interactions between circRNA and disease is time-consuming and expensive, effective computational methods are an urgent need for predicting potential circRNA–disease associations. In this study, we proposed a novel computational method named GATNNCDA, which combines Graph Attention Network (GAT) and multi-layer neural network (NN) to infer disease-related circRNAs. Specially, GATNNCDA first integrates disease semantic similarity, circRNA functional similarity and the respective Gaussian Interaction Profile (GIP) kernel similarities. The integrated similarities are used as initial node features, and then GAT is applied for further feature extraction in the heterogeneous circRNA–disease graph. Finally, the NN-based classifier is introduced for prediction. The results of fivefold cross validation demonstrated that GATNNCDA achieved an average AUC of 0.9613 and AUPR of 0.9433 on the CircR2Disease dataset, and outperformed other state-of-the-art methods.
1. Introduction
Circular RNAs (circRNAs) are a new class of endogenous non-coding RNA lacking a 5
′.cap and a 3
polyadenylated tail
[1][2]. Since circRNAs were first discovered, in the 1970s, they have been considered as splicing errors
[3][4]. In the past decade, with the development of high-throughput sequencing technology, a large number of circRNAs have been identified in mammalian cells
[5][6]. Researchers have found that circRNAs are widely expressed in human tissues, and have stable structure and tissue-specificity. The mechanism of circRNA expression remains unknown, and how the biogenesis of circRNA affects its unique regulatory pattern remains limited
[7]. Studies have revealed that many circRNAs perform their biological functions by acting as sponges of microRNA or RNA-binding proteins, by regulating protein function or by being translated themselves
[8][9][10].
Cumulative evidence has indicated that many circRNAs are involved in human diseases, especially cancers
[11]. For example, circHIPK3 has been found significantly up-regulated in colorectal cancer (CRC) tissues by sponging miR-7 to inhibit miR-7 activity
[12]. Hsa_circ_0000190 was down-regulated in gastric cancer (GC) tissues and plasma from patients with GC. Compared with common biomarkers such as CEA and CA19-9, it has better sensitivity and specificity, and can be used as a novel biomarker for diagnosis of gastric cancer
[13]. Researchers have identified that the expression of hsa_circ_0005075 is significantly different between hepatocellular carcinoma (HCC) and normal tissues
[14]. The expression of Hsa_circ_0001649 was significantly different between HCC and normal liver tissues
[15]. Moreover, circRNAs have also been related to other human diseases. CircANRIL is related to atherosclerotic disease by binding to pescadillo homolog 1 (PES1), which then impairs pre-rRAN processing and ribosomal biogenesis, results in the activation of p53, and thereby induces apoptosis and inhibits proliferation
[14]. Recent studies have shown that the circRNA level in the brain is associated with Alzheimer’s disease (AD)
[16]. Compared with the control group, Li et al. have found that 112 circRNAs were up-regulated and 51 circRNAs were down-regulated in AD patients
[17], which also were enriched in AD-related pathways, and the clinical guidance of circ-AXL, circ-GPHN and circ-PCCA in disease management of AD patients was identified.
As researchers have realized that circRNAs are abundant in mammalian cells, evolutionarily conserved and stable, and could serve as better biomarkers
[18], databases of rich circRNA information, such as circBase
[19], circ2traits
[20], CircFunBase
[21] have been built for study. Furthermore, researchers have also manually curated evidence from published literature, established databases such as circRNADisease
[19], CircR2Disease
[22], Circ2Disease
[23], and circAtlas
[24]. While experimental verification is expensive and time-consuming, computational methods have gradually introduced inferring potential circRNA–disease associations. Lei et al. first proposed a path weighted method to predict disease-related circRNAs. They calculated disease semantic similarity, disease functional similarity and integrated with the Gaussian Interaction Profile (GIP) kernel similarities. Then, they constructed a heterogeneous network and adopted the depth-first search (DFS) to traverse nodes in the network and calculate the predictive score
[25]. Yan et al. developed the DWNN-RLS method based on Regularized Least Squares of Kronecker product kernel for predicting circRNA–disease associations, and obtained AUC values of 0.8854, 0.9205 and 0.9701 in fivefold, 10-fold and leave-one-out cross validation, respectively
[26]. Another graph-based method KATZHCDA achieved the best AUC values of 0.7936 and 0.8469 in fivefold CV and LOOCV, respectively
[27]. Xiao et al. developed a weighted low-rank approximation optimization method with dual-manifold regulations to infer potential circRNA–disease associations
[28].
Deep learning algorithms have also been introduced in this field. Deepthi et al. proposed an ensemble method named AE-RF, which extracted features via deep autoencoder, and then used random forest for prediction. As a result, this method achieved 0.9486 and 0.9552 in fivefold and 10 fold CV, respectively
[29]. Li et al. used DeepWalk to extract node features in the circRNA–disease network, and used a network consistency projection algorithm for circRNA–disease interactions prediction
[30]. Wang et al. designed GCNCDA using FastGCN to extract high-level features, and by applying Forest PA classifier for prediction
[31]. As a result, it achieved an AUC value of 0.909 in fivefold CV based on circR2Disease dataset. Bian et al. developed GATCDA method based on graph attention network to obtain representation of circRNAs and diseases, calculated the probability score by dot production
[32], and yielded an AUC value of 0.9011.
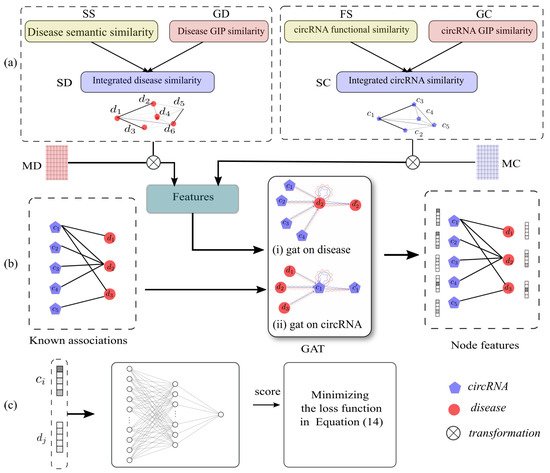
In this study, we proposed a novel computational method named GATNNCDA to predict potential circRNA–disease associations, based on graph attention network and multi-layer neural network. To be specific, GATNNCDA first integrates circRNA functional similarity, disease semantic similarity and the GIP similarities. Secondly, GATNNCDA utilizes linear transformation to project the integrated similarity matrices into the same space, and applies a graph attention network to extract dense representations of nodes in the heterogeneous circRNA–disease graph. Furthermore, a multi-layer neural network is constructed to infer the associations between circRNAs and diseases. The framework of GATNNCDA is shown in Figure 1. In summary, our contributions are listed as follows:
Figure 1. The framework of GATNNCDA. It consists of three steps: (a) similarity integration for circRNA and disease, (b) GAT-based feature extraction, and (c) NN-based classification.
-
We proposed an end-to-end framework for inferring disease-related circRNAs, which can effectively and accurately infer the potential associations between circRNAs and diseases.
-
We made use of GAT to extract low-dimensional dense representations of circRNAs and diseases, and these presentations had rich structural and semantic information of the heterogeneous circRNA–disease graph.
-
We proposed a NN-based classifier, and applied a sampling strategy to construct balanced samples. In addition, we designed cross-entropy loss with L2 regularization to make the training process fast and robust.
-
We demonstrated the predictive performance of our method by extensive experiments via fivefold cross validation and case studies, and achieved competitive results on CircR2Disease and circRNADisease datasets.
2. Case Studies
To further evaluate the prediction ability of our proposed method, we performed two case studies in this section. We trained GATNNCDA on CircR2Disease dataset
[22], and then verified the candidates on circRNADisease
[19] and circAtlas v2.0
[24] datasets. The first case study was conducted on breast cancer, which is one of the most common cancers in women. In particular, we constructed the positive samples with all known associations between circRNAs and diseases in the CircR2Disease. Meanwhile, we randomly chose the same number of negative samples from the unknown associations. Based on these training samples, we built the GATNNCDA and calculated the scores between breast cancer and each circRNA. Finally, we selected the top 20 related circRNAs for analysis. As shown in
Table 1. Top 20 predicted circRNAs related to Breast cancer based on circR2Disease dataset.
| Rank |
circRNA |
Evidence |
Rank |
circRNA |
Evidence |
| 1 |
hsa_circ_0007534 |
II |
11 |
hsa_circ_0068033 |
I; II |
| 2 |
hsa_circ_0011946 |
II |
12 |
circamotl1hsa_circ_0004214 |
I; II |
| 3 |
hsa_circ_0093859 |
II |
13 |
hsa_circ_0006528 |
I; II |
| 4 |
circrna-000911 |
II |
14 |
hsa_circ_0002874 |
I; II> |
| 5 |
circrna-001283 |
PMID:29431182 |
15 |
hsa_circ_0001667 |
I; II |
| 6 |
circrna-001175 |
II |
16 |
hsa_circ_0085495 |
I; II |
| 7 |
circrna-100438 |
PMID:29431182 |
17 |
hsa_circ_0086241 |
I; II |
| 8 |
hsa_circ_0001982 |
I; II |
18 |
hsa_circ_0092276 |
I; II |
| 9 |
hsa_circ_0001785 |
I |
19 |
hsa_circ_0003838 |
I; II |
| 10 |
hsa_circ_0108942 |
I; II |
20 |
circvrk1 |
I; II |
I, II denote circRNADisease, circAtlas v2.0.
The second case study is performed on hepatocellular carcinoma. It is the most common form of liver cancer, with a higher incidence in patients with long-term liver diseases
[33]. We utilized GATNNCDA to calculate the correlation score with circRNAs and then sorted by descending order. The top 20 hepatocellular carcinoma related cirRNAs are listed in
Table 2. We can see that 10 of the top 20 are verified by the validation datasets, and the other eight candidates have been conformed in relevant literature, e.g., hsa_circ_0000520 is one of the three circRNAs that showed significantly different expression levels in HCC tissues
[14]. Therefore, the unknown associations with high scores are likely to be correlated.
Table 2. Top 20 predicted circRNAs related to hepatocellular carcinoma based on circR2Disease dataset.
| Rank |
circRNA |
Evidence |
| 1 |
circc3p1 |
II |
| 2 |
hsa_circ_0067531 |
II |
| 3 |
circarsp91hsa_circ_0085154 |
II |
| 4 |
circmto1hsa_circrna_0007874hsa_circrna_104135 |
II |
| 5 |
hsa_circ_0005986 |
I; II |
| 6 |
hsa_circrna_100338circsnx27 |
PMID:28710406 |
| 7 |
hsa_circrna_104075 |
I; II |
| 8 |
hsa_circrna_102049 |
PMID:28710406 |
| 9 |
circrna_000839 |
II |
| 10 |
circzkscan1hsa_circ_0001727 |
I; II |
| 11 |
hsa_circ_0004018 |
I; II |
| 12 |
hsa_circ_0005075 |
II |
| 13 |
hsa_circrna_100571 |
PMID: 29609527 |
| 14 |
hsa_circrna_400031 |
PMID:29609527 |
| 15 |
hsa_circrna_102032 |
PMID: 29609527 |
| 16 |
hsa_circrna_103096 |
PMID:29609527 |
| 17 |
hsa_circrna_102347 |
PMID:29609527 |
| 18 |
hsa_circrna_000167hsa_circ_0000518 |
unknown |
| 19 |
hsa_circ_0000520 |
PMID:27258521 |
| 20 |
hsa_circ_0000172 |
unknown |
I, II denote circRNADisease, circAtlas v2.0.
3. Conclusions
Cumulative evidence has shown that circRNAs play an important role in progression of human diseases, and are suitable as promising disease biomarkers for prevention, diagnosis and treatment. As traditional biological identification is very costly and time-consuming, more and more computational methods have been introduced in this field. In this study, we proposed a novel computational method called GATNNCDA for predicting potential circRNA–disease associations. GATNNCDA achieved a better performance than other state-of-the-art methods by combining similarity integration, graph attention network and multi-layer neural network. In particular, we performed fivefold CV for evaluation, and obtained the best performance of AUC of 0.9742, AUPR of 0.9707. The average values of AUC and AUPR for under 50 experiments were 0.9613 and 0.9452. Furthermore, case studies on breast cancer and hepatocellular carcinoma have also demonstrated that GATNNCDA can be a useful tool for predicting potential disease-related circRNAs.
However, GATNNCDA still has some limitations. The initial node features may not be perfect. Recall that similarity integration as initial node representations would affect the final performance. Nonetheless, known interactions between circRNA–disease associations are insufficient. In addition, circRNA functional similarity and GIP similarity may be inaccurate. Therefore, more biological information such as circRNA–miRNA association or circRNA sequence will be used for further study to construct more accurate node features, especially for some unseen circRNAs. Furthermore, the NN-based classifier of GATNNCDA requires negative samples for training, which are rarely reported in the literature. Randomly sampling from the unknown associations in a CircR2Disease dataset would introduce bias. In the future, we will seek a better negative sampling strategy to promote the performance of GATNNCDA.