2. Discussion
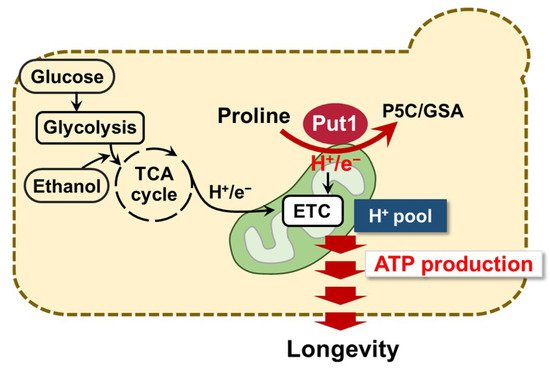
In this study, we discovered that the proline oxidase Put1 contributes to the chronological lifespan of yeast. Upon yeast cells entering the aging period, PUT1 expression was positively regulated by Put3 and was associated with longevity. More intriguingly, Put1 helped sustain mitochondrial membrane potential and ATP production, leading to maintenance of the energy metabolism. Our present data suggest a possible mechanism for the longevity regulation via proline metabolism (Figure 1). Yeast cells normally produce ATP from glucose and ethanol via glycolysis in the cytosol and/or ETC in mitochondria, respectively. When all glucose and ethanol are consumed, yeast cells probably uptake proline from the environment and start to utilize proline as an energy source via its oxidation by Put1. This conversion of proline into P5C results in the generation of electrons that are donated to ETC through FAD to generate ATP. The mechanism functions for the homeostasis of energy metabolism, which contributes to longevity.
Figure 1. Schematic model of longevity regulation by proline oxidation. In the absence of principal energy sources, such as glucose and ethanol, yeast cells start to utilize proline as an energy source via the Put1-catylatic reaction. This reaction results in the generation of electrons which are donated to ETC, which generates ATP. Such a mechanism may contribute to longevity.
Proline is a multifunctional amino acid in many organisms. In addition to being a proteogenic amino acid, proline functions as a stress protectant, namely an osmolyte, oxidative stress protectant, protein-folding chaperone, membrane stabilizer and scavenger of reactive oxygen species
[4][5][6][7]. Therefore, proline itself may protect yeast cells against various stresses caused by aging and regulate the longevity. However, our present study showed that the amount of proline in the cell does not affect the lifespan. In addition, proline addition had no effect on the lifespan of
put1Δ cells. These results indicated that the use of proline for ATP synthesis is more important for longevity than as a protective molecule. Aged cells are known to accumulate misfolded proteins or protein aggregates inside the cell due to downregulation of protein quality control systems
[21]. Molecular chaperones are central components of such systems and require ATP for protein refolding. Thus, ATP synthesized via proline oxidation catalyzed by Put1 may be used to support the activity of molecular chaperones during aging period, leading to removal of unfolded proteins and prolongation of the longevity. Further studies are needed to reveal that the proline catabolism helps the chaperone activity under aging.
Since yeast cells can utilize various nitrogen sources, the sensing of nitrogen sources is important for optimizing cellular metabolism. Generally, the presence of a preferred nitrogen source, such as ammonium ions or glutamine, represses the expression of catabolic genes compared to nonpreferred nitrogen sources, such as proline and leucine, through nitrogen catabolite repression (NCR)
[22][23][24]. All of the proline-metabolizing genes are NCR-regulated genes; that is, their transcription is repressed by the NCR repressors Ure2 and Dal80 when a preferred nitrogen source is present
[25]. In addition, the proline transporters Gap1 and Put4 are well-known to be subjected to post-translational regulation, which suppresses their transport activity by the ubiquitin ligase Rsp5
[26][27]. Thus, proline appears to be consumed, similar to other nonpreferred nitrogen sources. In fact, however, yeast cells rarely uptake and consume extracellular proline, even though they do use other nonpreferred nitrogen sources, such as leucine and 4-aminobutanoic acid
[28][29][30]. In short, both the uptake and metabolism of proline are strongly inhibited in cells compared with other nonpreferred nitrogen sources. In light of our present study, yeast cells may adopt a strategy of leaving proline outside the cell as a last resort for survival. The presence of energy sources such as glucose strongly inhibits the uptake of proline. In the absence of other energy sources, yeast cells may uptake proline via Gap1 and Put4 and oxidize proline by Put1, which produces ATP to assist in cellular survival. This suggests that proline metabolism is not only regulated by nitrogen sources but also by energy sources such as glucose. Our previous study showed that
PUT1 induction is positively regulated by the general stress transcription factors Msn2/4
[18]. The important fact is that Msn2/4 is activated by the low-glucose sensor Snf1
[31]. Hence, Snf1 may sense energy source depletion in the environment and subsequently Msn2/4 and Put3 may work cooperatively to activate proline metabolism, leading to maintenance of energy homeostasis. Further studies will be needed for understanding the detailed mechanism and physiological significance of proline-mediated energy homeostasis.
In this study, we found that proline taken up from outside the cell, rather than proline synthesized inside the cell, contributes to the longevity of yeast. On the other hand, a previous study indicated that the intracellular proline regulates the replicative lifespan of yeast
[18]. Generally, proline is present in the cytosol, but most of excess proline is stored in the vacuoles
[32]. Since a mechanism for active transports of proline from the vacuoles to the cytosol has not been identified, proline in the vacuoles may remain there indefinitely. The most severe stress under replicative events is considered to be osmotic stress, and vacuoles are important organelles for the adaptation to osmotic pressure
[33]. Thus, proline maintained in vacuoles may be functional during the replicative lifespan. In the case of regulating the chronological lifespan, however, proline must be transported from the cytosol to the mitochondria to serve as a substrate for Put1. Therefore, it is likely to be important for yeast cells to uptake extracellular proline upon entering the aging period.
Proline metabolisms is highly conserved across various organisms. In terms of proline degradation, the localization of this process and the properties of the enzymes involved are very similar between yeasts and mammals. It is known that the mitochondrial proline oxidase is implicated in supporting ATP production, protein synthesis, and redox homeostasis in cancer cells
[34]. One of the most lethal capacities of cancer cells is their ability to metastasize to distant organs. Recently, Elia et al. suggested that proline metabolism supports the metastatic cascade via the production of ATP
[35]. Importantly, cancer cells in metastatic tissue exhibit higher expression of proline oxidase than normal cells. Accordingly, inhibition of proline oxidase activity impairs metastasis formation in different kinds of metastatic breast cancer
[35]. However, the details of how ATP production is involved in metastatic progression are still unknown. Considering our present data, proline oxidase may function as an enzyme to prevent cellular senescence mediated by ATP production. Likewise, inhibition of senescence is well-known to induce cancer formation and metastasis
[36]. Thus, an inhibitor of proline oxidase would be a promising candidate as a drug that interferes with the metastatic cascade. Yeast cells can grow on the medium containing proline as a sole nitrogen source, but Put1, an orthologue of proline oxidase, is essential for such growth. Therefore, inhibitors of proline oxidase may be screened by a chemical biology-based study utilizing yeast.
The aging process is a part of the life cycle of all organisms. Yeasts are very important microorganisms for various industrial fields, such as the production of fermented foods and useful compounds. A high fermentative capacity is a key factor for their biotechnological application. Since the yeast performance in biotechnological applications is dependent on levels of cell viability and vitality, the extension of the lifespan is important to maximize these processes. Hence, isolating and/or engineering yeast strains with the appropriate localization and concentration of proline could contribute to improvements in fermentation and other kinds of biochemical productivity.