1.1. Currently Known Pathophysiology, Molecular Diagnosis and Treatment Strategies in CLL
CLL is the most frequent lymphoproliferative disease in the western world
[1][2] characterized by the clonal proliferation and progressive accumulation of mature, typically CD5-positive B-cells in the blood, bone marrow, and secondary lymphoid tissues
[2][3][4]. It shows a high biological, genetical, molecular and clinical diversity
[1][5], projecting its highly heterogenous pathophysiology (
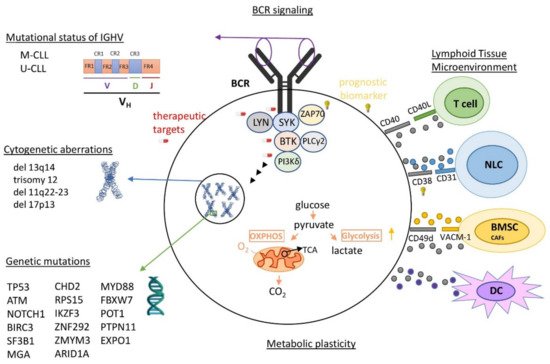
Figure 1). Among the known features with clinical relevance in the pathobiology of CLL are the highly genetic mutations acting either independently or in combination with chromosomal rearrangements
[1][5]. Driver mutations have been associated with adverse clinical outcomes, and thus serve as biomarkers, indicators of therapeutic options or as potential therapeutic targets
[2][4][5][6][7][8]. Somatic mutations in immunoglobulin heavy chain variable region gene (IGHV), activating B cell receptor (BCR)-signaling kinases lead to the lower survival and proliferation of CLL cells, providing patients with “mutated” M-CLL, which is a better clinical outcome vs. “unmutated” U-CLL patients
[2][9]. It is important to mention that the signaling of UM-CLL is generally highly responsive to the antigenic stimulus, while M-CLL are anergic. Continual or repetitive BCR signaling adds further complexity in CLL pathogenesis, contributing to autophagy regulation, promoting tumor survival, proliferation, and consequently tumor progression
[10]. Complex karyotype (CK), defined by the presence of at least three genetic abnormalities in the same clone, is detectable in 14–34% of CLL cases and it is recommended as a new negative prognostic biomarker associated with an adverse outcome and worse response to chemoimmunotherapy
[11][12][13][14][15]. Other intriguing features of vital significance in the growth, survival, and drug resistance of CLL cells are metabolic plasticity and signals from the lymphoid tissue microenvironment (LTME)
[2][4]. Metabolic plasticity involves the main metabolic pathways of mitochondrial biogenesis and bioenergetics, ROS production, and adaptation to intrinsic oxidative stress, found to be elevated in CLL
[16]. LTME produces various essential proteins and metabolites
[3][17] modulating the redox and metabolic state of CLL cells
[18] and switching either to oxidative phosphorylation (OXPHOS) or glycolysis
[19]. Furthermore, enhanced BCR signaling induces the metabolic activation of CLL cells through OXPHOS, energetically supporting the transcription and translation processes
[20].
Figure 1. Currently known heterogeneity in the pathophysiology of CLL. Some of the most important elements of CLL pathophysiology are: (1) highly varying genomic mutations, (2) loss or addition of large amounts of chromosomal material, (3) mutational status of variable region of IGHV, (4) frequent activation of BCR signaling and (5) continuous proliferating signals from the cancerous microenvironment. IGHV: immunoglobulin heavy chain variable region genes; M-CLL: mutated CLL; U-CLL: unmutated CLL; BCR: B cell receptor; NLC: nurse-like cells; BMSC: bone marrow stromal cells; DC: dendritic cells; OXPHOS: oxidative phosphorylation; TCA: citric acid cycle.
The molecular diagnostic criteria in CLL guidelines and beyond traditional Rai or Binet staging
[21] include (i) the co-expression of CD5 with the B-cell antigens CD19 and CD20, (ii) characteristically lower levels of surface immunoglobulin, CD20, and CD79b (vs. normal B cells), (iii) the expression of kappa or lambda immunoglobulin (Ig) light chains
[3][22] and (iv) the identification of specific gene mutations and serum markers
[2][3][22][23]. Additionally, the CLL International Prognostic Index (CLL-IPI) proposes a weighted grading of five parameters: (i)
TP53 dysfunction, (ii) mutational status of IGHV, (iii) serum level of β2-microglobulin, (iv) clinical stage, and (v) age
[3]. Furthermore, an increasing number of studies are supporting the use of new biomarkers for the diagnosis, prognosis of clinical course and therapeutic decision, such as newly approved driver genes
[5][24][25][26][27][28][29][30] serum micro-RNAs
[31], etc. Interestingly, assessment of the minimal residual disease (MRD), referring to the small numbers of CLL cells that remain in patients in remission during or after treatment, is an emerging prognostic biomarker of progression-free and overall survival
[23][32].
A plethora of pharmacological targets have been investigated in CLL. Patients, according to their clinical history, are prioritized to therapeutic options, including chemotherapy, immunotherapy (IT), chimeric antigen receptor and other targeted therapeutic strategies, used alone or in combination. More specifically, chemotherapeutic agents are still used in many cases as a first-line treatment.
[2][3]. In chemo-immunotherapy (CIT), mAbs bind in the surface antigens of CLL cells, resulting in apoptosis, complement-dependent cytotoxicity (CDC) or antibody-dependent cellular cytotoxicity (ADCC). Different combinations of several agents have been reported and evaluated in several publications
[2][3][22][33][34]. Inhibitors targeting the aberrantly regulated components of apoptosis, and of BCR signaling in CLL
[2][3][22][33][35][36], have started to replace CIT, in first- and second-line indications
[3][36] and many other new generations or under investigation agents
[36][37][38][39]. The therapy using CAR-T cells represents a recent therapeutic option for some CLL patients
[40][41]. Finally, among the promising CLL therapies under investigation targeting several deregulated pathways are the cross-talk between CLL cells, the tumor microenvironment
[42][43], the Wnt signaling pathway
[44], various miRNAs
[45], the Notch2 signaling pathway
[46], the mitochondrial metabolism
[16] and the epigenetic modifications
[47].