1000/1000
Hot
Most Recent

Despite constant progress in understanding its pathology and associated therapeutic actions by targeting lifestyle changes and novel drug treatment strategies, arterial hypertension currently represents one of the major causes of cardiovascular morbidity and mortality in Europe, with severe potential complications. Significant distinct individual variations of responses to antihypertensive therapy suggest that genetic analysis may provide new important data regarding an accurate definition of prognosis and, most significantly, an adequate choice of treatment, therefore preventing potential complications. Data regarding the multifactorial genetic inheritance of essential arterial hypertension have long been considered; however, given the current rise of genetics and genomics, DNA sequencing could represent a step toward specific genetic variation-based therapy in hypertensive patients with uncontrolled blood pressure, despite standard antihypertensive treatment.
Arterial hypertension currently represents the main risk factor responsible for global mortality and burden of disease, with a strong correlation between high blood pressure and cardiovascular disease [1][2].
Thus, a better understanding of the genetic basis of a polygenic disease such as hypertension might bring antihypertensive treatment one step closer to reaching the recommended targeted values of therapy. Likewise, a genetic profile could be helpful in performing individual risk and prognosis stratifications which are based on the identification of specific genetic variants.
Since an adequate definition of expressed sequences in the human genome represents a useful instrument for defining gene families, proteins and primary patterns of expression in tissues and in specific diseases, a targeted genetic analysis for hypertensive patients would allow for the identification of genes responsible for the disease and an accurate characterization of genomic regions that are linked to arterial hypertension. The study of genetic variants has evolved in the direction of pharmacogenomics: genomic variation inducing individual responses to therapy. This concept can be useful in redefining antihypertensive pharmacological therapeutic actions, as well as for understanding the reason why some individuals respond to standard treatment and others do not.
Certain factors seem to be connected to the genetic individual variability of a response to antihypertensive treatment: different metabolic profiles and genetic variations of metabolizing enzymes, the genetic variability of sodium sensitivity and proteins from renal tubules (responsible for the regulation of ion transport or variability of response to diuretics) (Table 1).
Table 1. Factors influencing genetic variability of response to antihypertensive therapy.
| Factors Influencing Genetic Variability of Response to Antihypertensive Therapy | |
|---|---|
| 1 | Genetic variations of metabolizing enzymes |
| 2 | Genetic variability of sodium sensitivity |
| 3 | Genetic variability of proteins regulating renal tubules ion transport |
Concerning etiology, most antihypertensive patients lack an obvious cause and are diagnosed with essential arterial hypertension, a heterogeneous disease with important genetic implications. From this point of view, the Genome Wide Association Study (GWAS) has been an important tool for identifying the genetic implications of essential arterial hypertension [3]. A GWAS provided the opportunity to type, at a large scale, a multitude of Single Nucleotide Polymorphisms (SNP) and, in the setting of large consortia, many studies were published that allowed for the identification of over 100 SNPs that are linked to high levels of blood pressure (see Table 2) [4][5][6][7][8][9].
Table 2. Polygenic form of essential arterial hypertension, data from [9].
| BP Implication | Genes Involved | References |
|---|---|---|
| Systolic BP | ATP2B1, CYP17A1, PLEKHA7, SH2B3 | Levy et al., 2009 [4] |
| Diastolic BP | ATP2B1, CACNB2, CSK-ULK3, SH2B3, TBX3-TBX5, ULK4 | Levy et al., 2009 [4] |
| Systolic or diastolic BP | CYP17A1, CYP1A2, FGF5, SH2B3, MTHFR, c10orf107, PLCD3 | Newton-Cheh et al., 2009 [10] |
| Pulse pressure | CHIC2/PDGFRA, PIK3CG, NOV, ADAMTS8 | Wain et al., 2011 [11] |
| Mean arterial pressure | CHIC2/PDGFRA, PIK3CG, NOV, ADAMTS8 | Wain et al., 2011 [11] |
BP-blood pressure.
At first, identifying the cause of essential hypertension remained a difficult task. Using the classic GWAS approach and the “common disease–common variant” hypothesis, genetic variations that could only be responsible for about 3% of essential arterial hypertension genetic causes were highlighted [3]. Thus, the idea that variants too rare to be identified by the GWAS, as well as those which could explain missing information, emerged. The use of new techniques such as systemic genetic approaches, related statistical methods, analyzing intermediate phenotype, transcript, protein and metabolite levels represented an important step in identifying rare genetic causes of essential hypertension. [12] In this field, Exome Chips Based studies and Next generation sequencing are useful tools in underlying rare variants, genes and pathways responsible for inducing essential hypertension. These studies were conducted after the 1000 Genomes Project became available, and they allowed for the development of array-based genotyping platforms (such as Illumina, San Diego, CA, USA) that provided the opportunity to identify a much greater number of nucleotide variabilities in comparison to GWAS [13][14][15][16][17][18][19][20][21][22][23].
One important inquiry regarding current antihypertensive treatment is whether a gender-related differentiation of this therapeutic approach should be considered in daily medical practice. This judgement is based on several facts. The first is that, despite current gender-neutral recommendations, it is a well-known fact that cardiovascular risks are different in female patients than they are in male patients, with data reporting a higher prevalence for worse risk factor profiles for female patients [24][25][26][27]. Among the factors that may influence this trend are advanced onset age of coronary heart disease in women and delayed medical diagnosis due to a lack of awareness from both health care professionals and female patients concerning coronary heart disease in women.
Data from the EUROASPIRE V European survey established that, related to cardiovascular risk factors control, women were less likely to perform an adequate level of physical activity and to achieve targeted values for glycated hemoglobin (HbA1c) and low-density lipoprotein cholesterol (LDL-C). Additionally, female patients were more likely to be characterized by obesity, non-smoking and with a medical history of diabetes [25][26][27]. These data are consistent with findings from other studies: the VIRGO (Variation in Recovery: Role of Gender on Outcomes of Young AMI Patients) study included more than 3,500 acute coronary syndrome patients and revealed a higher prevalence for diabetes and obesity in women, and the SWEDEHEART (Swedish Web System for Enhancement and Development of Evidence-Based Care in the Heart Disease Evaluated According to Recommended Therapies) registry evaluated 51,620 coronary patients who reported gender differences regarding blood pressure control and LDL-C targets, with a worsening prognosis for women [28][29]. Similar results were demonstrated by the REACH Registry after analyzing patients with documented arterial disease; analysis revealed that women were more likely to be obese and have uncontrolled cholesterol levels [30].
Beyond cardiovascular risk profiles, a gender-related individualization of antihypertensive therapeutic approaches is equally significant. Vynckier et al. reported that, related to antihypertensive treatment, no gender differences were observed for treated hypertension (46.3% vs. 46.6%; p = 0.43). Additionally, their results established that women were less likely to have uncontrolled hypertension (30.0% vs. 23.5%; p = 0.004), even though they were also more likely to use antihypertensive medication [25].
All these data underline the fact that the prevalence, awareness and, most importantly, the treatment of hypertension is variable among male and female patients.
One other possible clarification is represented by the fact that the mechanisms responsible for this pathology may be diverse and closely related to a gender factor, such as the renin–angiotensin system, sympathetic nervous activity, endothelin-1, sex hormones, and the immune system. Additionally, particular attention should be paid to the key role of aldosterone in influencing cardiovascular risk and immune system activation, with data emphasizing that the efficacy of mineralocorticoid receptor antagonists depends on aldosterone values, indicating a genetic difference in sensitivity to aldosterone [31]. In addition, Te Riet et al. demonstrate that high aldosterone levels in combination with high salt intake may contribute to increased endothelial dysfunction and lead to a rise blood pressure, independent of the renal effects of aldosterone [32].
The prevalence of hypertension is higher in men compared to women, at least in young patients under the age of 50. Beyond this age, the prevalence of hypertension is greater among women, with menopause and hormonal imbalances, as well as the activation of the renin–angiotensin system (RAS) and sympathetic and immune systems, contributing to increased blood pressure values [33][34][35][36] (Figure 1).
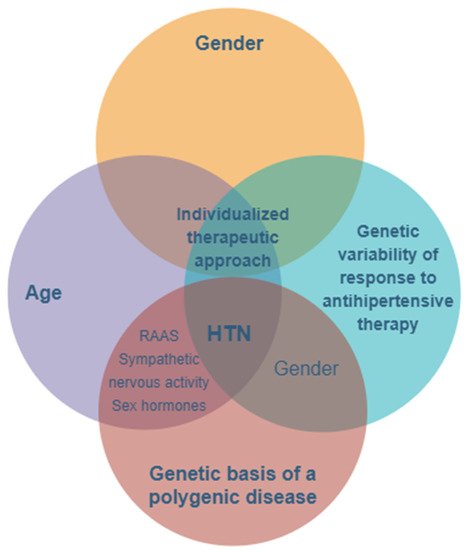
Figure 1. The main features and key factors for personalizing treatment in hypertension with a genetic- and gender-based substrate. HTN—hypertension; RAAS—renin–angiotensin–aldosterone system.
Last but not least, a recent study by Hongwei et al. indicates that related to specific blood pressure values, lower values of systolic blood pressure are associated with increased cardiovascular risk in women compared to men. They add that the risk of acute coronary syndromes for women with values of systolic blood pressure (SBP) between 110 and 119 mm Hg was comparable to the myocardial infarction risk for men with SBP ≥160 mm Hg [37].
The identification of susceptibility genes involved in multifactorial hypertension is based on linkage and gene association studies, which aim to establish a significant association of a certain chromosomal region or an allelic variant with the disease and are based on the hypothesis of “common disease–common variant”; this hypothesis starts from the premise that the main alleles of susceptibility can be identified in all patients with the same condition [38].
Chain assays were performed in families of patients based on the assumption that patients (usually affected siblings) will more commonly share a certain allele (of a candidate gene) compared to healthy people in the same family. Association studies are performed in a population to compare the incidence of a certain gene polymorphism with its incidence in a control group; if a specific allele of the polymorphic locus has a significantly higher presence in individuals with the disease compared to those unaffected, then it can be assumed that the allele is involved in the pathogenesis of the disease.
Despite the complexity of the disease and its technical difficulties, several gene variants that confer susceptibility to hypertension were identified in the “pregenomic” period, especially genes encoding various components of the renin–angiotensin system, ion channels, or enzymes involved in the synthesis of aldosterone. The following were identified: the ADD1 gene (α-bringing, hydrostatic pressure change sensor, located on chromosome 4p16); the AGT gene (for angiotensinogen, located on chromosome 1q42-q43); the REN gene (for renin, located on chromosome 1q32); the ACE gene (for the angiotensin converting enzyme located on chromosome 17q23); the AT1R gene (for the angiotensin receptor located on chromosome 3q21); the GJA5 gene (for connexin 40, from the junctional structures of endothelial cells in the efferent arteriole and the juxtaglomerular apparatus) for the beta subunit of the non-voltage-dependent sodium channel type 1, located on the 16p13-p12 chromosome); and the CYP11B2 gene (for aldosterone synthesis, located on chromosome 8q21) [38].
In recent years, “case–control” association studies (based on the “common disease—common variant” hypothesis) have been conducted at the genome-wide level (GWAS), accelerating the decipherment of the genetic architecture of BP and HTN. Using genotyping with “high throughput” techniques (SNPs-type DNA microgrids/microchips), about one million genetic markers covering the entire genome can be investigated in a single scan of the human genome. Genotyping of tens of thousands of patients with the same disease and a control group of healthy individuals allowed for the identification of genomic regions that were significantly associated with the disease; these are regions in which variants of disease susceptibility genes are located. The discovery of the causal gene variant (in the identified chromosomal region) and the determination of the mechanism by which this variant participates in the development of the disease obviously require other studies, such as the following: fine mapping to minimize the size of the region in which the susceptibility gene could be found; analysis of DNA sequences in the determined minimum interval to identify the present gene/genes, each with numerous allelic variants; and functional testing of candidate gene variants to experimentally determine the functional effect of the identified gene and its role in disease pathogenesis [39].
From 2008 to 2011, seven GWAS studies for BP and/or hypertension were performed on large populations of different ethnic origins [40][39]. In total, 41 SNPs significantly associated with BP and/or hypertension were identified. In the chromosomal regions (≈100 Kb) or loci defined by these “signals”, the identification of some causal genes proved to be difficult (see Table 2), and only a few genes (NPPA, NOS3, UMOD) were clearly associated with a particular SNP located in the sequence of a gene. Most SNPs were located in regions rich in genes, and then, the “nearest gene” was taken into account (see table). At other times, SNPs involved “regulatory sequences” (binding sites for transcription factors or microRNA molecules) for more distant and yet-to-be identified risk genes.
High blood pressure has been shown to be more difficult to decipher than its significant heritability might have suggested. Many identified susceptibility genes have new functions in known pathogenic pathways or are likely to intervene in new pathogenic pathways. The elucidation of these aspects requires further studies but will certainly lead to improving the etiological diagnosis, establishing an individual profile of the disease and optimizing therapy by finding “customized solutions”. In support of this idea, we will refer, in conclusion, to the pharmacogenomics of antihypertensive drugs.
Despite the existence of many classes of effective drugs and many types of drugs in each class, the rates of BP control among hypertensives are disappointing (below 35%), which is mainly due to their low efficacy in some people. In addition, the therapeutic responses to a drug or a combination of drugs differ greatly between patients, as they are highly variable [41][42]. The solution to this drawback is to individualize therapy based on personal genetic information; in short, the pharmacogenomics of hypertension. To date, polymorphisms have been identified in several genes involved in the action of some drugs, which explain the individual variations of their effects: the ACE gene (encodes the angiotensinogen) and the angiotensin inhibitors; the ADD1 gene (encodes α-aducine, hydrostatic pressure sensor) and thiazide diuretics; the ADRB1 gene (encodes the adrenergic receptor b1) and beta-blockers; or the KCNMB1 gene (encodes the beta subunit of the calcium-activated potassium channel) and anti-calcium or beta-blocker drugs. However, the clinical potential of hypertension pharmacogenomics will need to be clearer and more accurate for practitioners to benefit from a rapid test that provides information on the use of first-line antihypertensive drugs. It is estimated that these goals will be achieved by 2022.