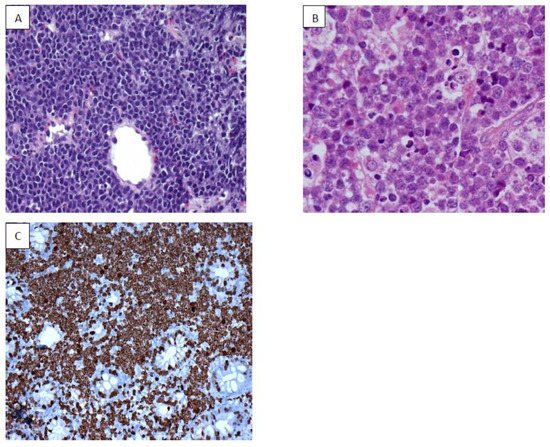
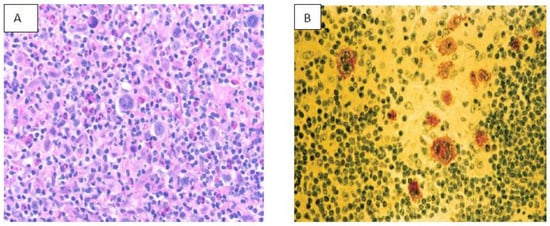
9. NK/T Cell Lymphoma (NKTCL)
EBV is well documented as an oncogenic virus in B cell neoplasm; however, its role in the pathogenesis of NKTCL has been complex. Mature NKTCL is a group of heterogeneous tumors that have complex treatment options, aggressive clinical presentation, and poor prognosis
[45]. The geographic distribution of EBV-induced NKTCL has been more concentrated in Asia (China, Korea), South America (Peru, Argentina), Central America (Mexico), and Central Africa. EBV in NKTCL is classified into nodal and extra-nodal types. The role of EBV in angioimmunoblastic T-cell lymphoma, nasal type of extranodal NKTL, and aggressive NK- cell leukemia is well researched and documented
[46].
The role of EBV infection in NKTCL has been hypothesized during the targeted killing of EBV-infected cells by NK/T cells. The patients with overlapping evidence of EBV- associated diseases are shown to have circulating EBV-infected NK/T cells (EBV positive NK/T cells)
[46][47]. This positivity is due to the effect of the “trogocytosis” phenomenon caused by glycoproteins (gp 350, gp 42, and gp85) and cellular protein (CD 21). This further induces the expression of EBNA (types 1,2,3A,3B,3C) and LMP (1,2A,2B) proteins that result in genetic polymorphisims. LMP1 acts as an active member of tumor necrosis factor (TNF) and downregulates NF-Kb and MAPK signal pathways
[45][46][48]. This further causes variation in the C-terminus of LMP1, deletion of 30bp, 33bp repeats, insertion of 15bp, and other amino acid substitutions. These genetic expressions, deletions, and variations have been studied in numerous molecular genetic pathways using whole-genome sequencing, targeted and exome sequencing. This has allowed the discovery of more target-specific therapies to these sets of complex malignancies
[46][49]. The most common genetic alteration in EBV-induced NKTCL is the loss of 6q21 (20–43% cases) followed by recurrent losses in chromosomes 1p4 and 5p13. JAK-STAT signaling pathways along with STAT3, KMTZD, and TP53 are the most recurrently mutated genes in EBV-induced NKTCL
[50].
Extranodal NKTCL occurs predominantly in midline facial structures and can also affect paranasal sinuses, orbit, jaw, and salivary glands. Symptoms vary from systemic features (fever, weight loss, night sweats) to hemophagocytic disorders
[51]. Imaging studies (CT, MRI, PET/CT) coupled with EBV-DNA biomarker assessment are essential for diagnosis
[52].
Treatment modality varies on the stage of NKTCL. Stage I/II are treated mainly with radiotherapy. Previous chemotherapeutic regimes using anthracycline-based CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) have shown to cause more toxicity and drug resistance
[53]. The recently introduced LVP (L-asparaginase, vincristine, and prednisone) and GELOX (gemcitabine, oxaliplatin, and L-asparaginase) are shown to be more effective in this stage
[51][54]. Advanced/relapsed/recurrent NKTCL are treated with selective chemotherapy but due to poor host response and drug-induced toxicity has resulted in poor patient prognosis in many of the cases. Hematopoietic stem cell transplant and immunotherapy are still under trial
[55].