1000/1000
Hot
Most Recent

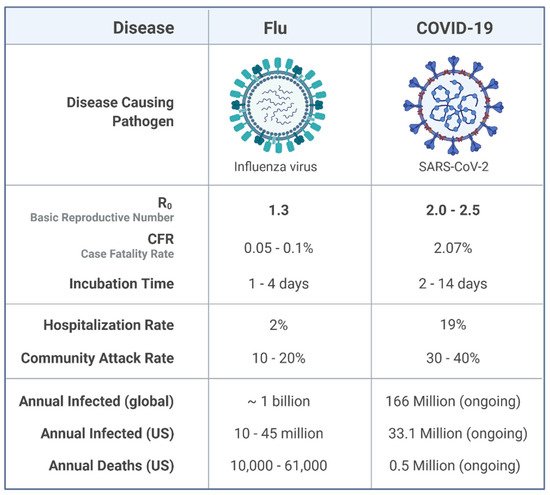
Influenza is a highly known contagious viral infection that has been responsible for the death of many people in history with pandemics. These pandemics have been occurring every 10 to 30 years in the last century. The most recent global pandemic prior to COVID-19 was the 2009 influenza A (H1N1) pandemic. A decade ago, the H1N1 virus caused 12,500 deaths in just 19 months globally. Now, again, the world has been challenged with another pandemic. COVID-19 and influenza viruses have very similar signs, and symptoms may explain the similar origin. According to a recent World Health Organization survey, the COVID-19 attack and disease burden in children have been much lower than influenza outbreaks, and the secondary household attack rate has also been low. This is in stark contrast to reports of the virus spreading quickly in enclosed spaces like hospitals or cruise ships, as well as a high prevalence of healthcare-associated infections.
| Gender proportion of COVID-19 cases | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 47.5 | 50 | 52.8 | 52.3 | 51.8 | 53 | 49.7 | 39.2 | |
| Female (%) | 52.5 | 50 | 47.2 | 47.7 | 48.2 | 47 | 50.3 | 60.8 | |
| Gender proportion of COVID-19 deaths | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 65 | 63.9 | 65.5 | 64.6 | 67.1 | 64.22 | 60.7 | 46.9 | |
| Female (%) | 35 | 36.1 | 34.5 | 35.4 | 32.9 | 35.8 | 39.3 | 53.1 | |
| Gender proportion of COVID-19 morbidity | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 0.94 | 3.17 | 4.16 | 3.38 | 3.32 | 3.02 | 2.55 | 2.85 | |
| Female (%) | 1.08 | 3.22 | 3.67 | 2.93 | 2.84 | 2.36 | 2.04 | 2.56 |
| Feature | COVID-19 | Influenza A(H1N1) | References | ||
|---|---|---|---|---|---|
| Epidemiology and Transmission | Fecal-oral transmission | Proved | Not proved | [39][50] | |
| Age composition | Most patients were older than 50 | Most patients were younger than 60 | |||
| Transmission mode | Asymptomatic/ symptomatic |
Symptomatic | [64] | ||
| Reproduction number | 3 | 1.5 | [52] | ||
| Incubation period | 4.9 | 1.4 | [51] | ||
| Treatment | Anti-viral drug | N3/ebselen/Remdesivir | Oseltamivir and zanamivir | [60][65][66] | |
| CP therapy | * | * | [67][68] | ||
| Vitamin D | * | * | [57][58] | ||
| MSC therapy | * | Not effective | [69] | ||
| Diagnosis | CRISPR-based SHERLOCK technique | * | Not develop | [70] | |
| qPCR | * | * | [71] | ||
| Gut microbiome | * | * | [72] | ||
| Lymphopenia | * | * | [73] | ||
| CT scan | Ground-glass opacities have frequently been placed in the periphery of lower lobes | Ground-glass opacities has a central, peripheral, or random distribution | [74] | ||
| Clinical manifestation | Acute lung injury | * | * | [44] | |
| Cardiovascular | * | * | [38] | ||
| Gastrointestinal | diarrhea | * | * | [75] | |
| nausea | * | * | |||
| vomiting | * | * | |||
| Molecular biology | Receptor for virus-cell entrance | ACE2 | Sialic acid receptor | [76][77] | |
| Genetic material | Just one positive-sense single-stranded RNA | Eight negative-sense single-stranded RNA | [78] | ||
| Location of replication | DMV (cytoplasm) | Nucleus | [19] | ||