Nanobiosensors are sensors in which various nanosize materials are used, however, large equipment is still required for whole sensing systems, because they are not, by themselves, able to detect nanoscale events. The current state-of-the-art nanobiosensors are based on several diagnostic methods, such as enzyme-linked immunosorbent assay (ELISA), flow cytometric immunoassay, electrochemical (amperometric, voltammetric and impedance), immunoassay, mass spectrometric immunoassay, chromatographic immunoassay, and different optical immunoassays. Contemporary multiplexed nanobiosensors are developed for the detection and quantification of clinically relevant analytes in biological and clinical samples (i.e., blood, urine, and saliva).
1. Introduction
The clinical diagnosis of many diseases depends on the accurate and unambiguous detection of various biomarkers, which may include proteins or other biomolecules
[1][2][3]. Immunoassays are a well-established tool for measuring analytes that are normally present at very low concentrations and cannot be determined accurately by other less expensive tests. Standard immunoassays are used to detect one specific analyte, however, for complex diseases such as cancer, diabetes, rheumatoid arthritis (RA) and osteoarthritis (OA), which involve a multitude of biomarkers, one analyte is not enough to obtain an early and accurate diagnosis. In this case, multiple immunoassays can be employed. However, immunoassays require samples that are collected, labelled, archived, and bio-banked according to established laboratory protocols before even conducting the assays, which makes this method challenging and time-consuming. In order to diagnose at the earliest stages of disease development, it is crucial to detect as much information as possible from small quantities of clinical samples. In this context, multiplexed immunoassays are an obvious and attractive approach, especially when sample volumes are limited. The main advantage of multiplexed analysis is the capability to detect multiple biomarkers qualitatively or quantitatively in a single sample. There are additional advantages to this approach, such as more data points from single samples, reduced cost per data point, fewer errors due to fewer samples, and increased throughput (). Conversely, cross-reactivity remains a significant challenge. If antibodies cannot distinguish between the analyte and other structurally similar components, the specificity of immunoreactivity drastically decreases, and as a consequence, multiplexed detection may not work properly in complex biological and clinical samples.
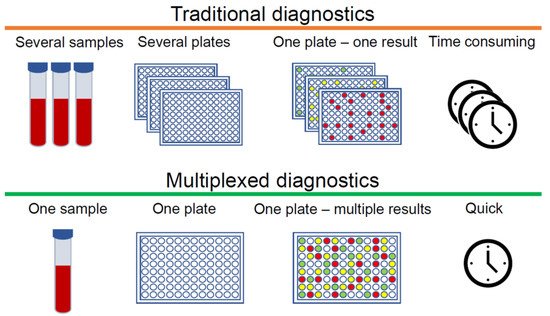
Figure 1. Differences between traditional and multiplexed diagnostics.
2. Quantum Dots
Since their first description in a biological context
[4] quantum dots (QDs) have been considered one of the most attractive luminescence label in biomedicine. Quantum dots are inorganic semiconductor crystals with a diameter of 1 nm to 10 nm (typically 2–6 nm), with optical properties which arise from interactions between electrons, holes, and their local environments
[5][6]. QDs absorb photons whose energy exceeds the photoluminescence excitation bandgap. In this process, electrons are transferred from the valence band to the conduction band. Due to many electronic states, QD photoluminescence excitation spectra are wide, but on the other hand, QD emission bands are narrow and symmetrical. These unique properties allow different QDs to be excited by the same light source and obtain different coloured visualisations in one biological sample. For comparison, the majority of organic fluorescent dyes have relatively narrow excitation bands and wide fluorescent band. Therefore, in order to perform multicolour imaging, several different light sources are required
[6]. By varying the size and composition of QDs, the emission wavelength can be tuned from blue to near-infrared
[6]. Additionally, because the surface of QDs are easily modifiable, it is easy to attach various antibodies and other biomolecules to them
[7]. Thus, QDs are highly applicable for multiplexed immunoassays. Here, we briefly review how QDs could be used for various multicolour biosensing applications.
2.1. QLISA
Quantum dot-linked immunosorbent assay (QLISA) is an approach similar to the enzyme-linked immunosorbent assay (ELISA), except QDs are used instead of enzymes
[8]. The principle of these two methods is the same. For both methods, the first step is antibody coating—the specific capture antibody is immobilised on high protein-binding plates. When the capture antibody is immobilised, the uncovered space on the plate is blocked with an irrelevant blocking protein, e.g., serum albumin. After that, samples and standard dilutions are added to the wells. In this step, the analyte adheres to the capture antibodies. In some ELISA cases, analytes can be attached directly on the plate surface, without using a capture antibody. The excess sample is removed, and then specific biotinylated detection antibody is added to the wells to enable detection of the captured protein. In the case of QLISA, QDs with conjugated detection antibody are used. After detection, the antibody binds to the analyte, and streptavidin conjugated with alkaline phosphatase/horseradish peroxidase (ELISA) or streptavidinated QDs (QLISA) is added to the wells which binds to the biotinylated antibody. For the ELISA method one addition step is required before analysis—addition of colorimetric substrate which form a coloured solution when catalysed by the enzyme. Samples are then analysed with microplate readers. The optical density in each well of the multi-well plate is measured, whereas for QLISA, photoluminescence of the QDs is registered.
ELISA is an established standard biomarker detection method that is widely used in biomedical research and in the clinical diagnostic setting
[2][9], whereas QLISA is still in the development stage. Nevertheless, the ELISA method still has some important disadvantages, which hopefully QLISA could solve. The main drawback of the conventional ELISA method is that the detection limit is barely less than the nanomolar concentration level, which is usually not enough to reach the clinical threshold of many protein biomarkers, especially in the early stages of disease
[10]. This disadvantage is due to low signal-to-noise ratio and limited signal amplification
[11]. An additional drawback is that traditional ELISA detects only one analyte in a sample, and multiplexed detections require several measurements simultaneously carried out
[12]. These disadvantages can be solved by using luminescent QDs instead of enzymatic colorimetric reactions for identification of the detection antibody. First of all, photoluminescence-based assays are more sensitive than absorbance-based assays
[13], thus the signal-to-noise ratio increases by measuring photoluminescence instead of absorption. Moreover, QDs have a high photoluminescence quantum yield, which allows acquisition of high signal intensities from relatively low concentrations
[14]. Therefore, QDs could work as amplifiers when low concentrations of analyte need to be detected. Finally, QDs, due to their optical and chemical properties, are good candidates for use in multiplexing ().
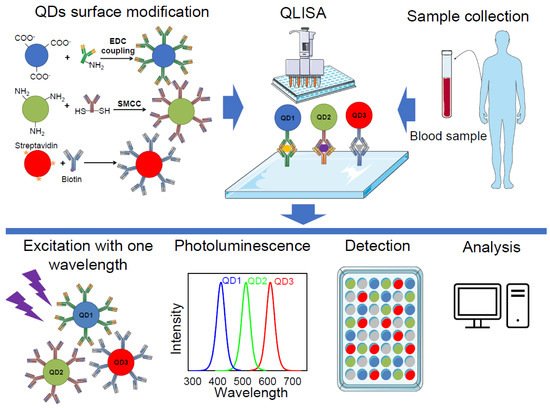
Figure 2. Multiplexed diagnostics using quantum dot-linked immunosorbent assay (QLISA). Firstly, the surface of the quantum dots (QDs) is modified with specific antibodies using 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) coupling, succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC) conjugation, or streptavidin–biotin interaction. Clinical samples from patients are processed and loaded to a QLISA plate, which is coated with capture antibodies. Then, modified QDs are loaded and the plate is scanned using a microplate reader. Different QDs in the sample are excited with the same wavelength and several bands of photoluminescence are detected. The final step is the analysis of the acquired data.
Mansur et al. (2015) successfully demonstrated the detection of the cluster of differentiation 20 (CD20) antigen, which is a biomarker overexpressed by B-lymphocyte cancer cells, using QLISA
[15]. Suzuki et al. (2017) reported the application of QLISA for the detection of the multifunctional cytokine interleukin 6 (IL-6) which is involved in many cellular processes, such as receptor synthesis, inflammation, cell proliferation, and cancer cell signalling
[8]. Functionalised QDs were used in QLISA to detect and quantify IL-6. Results demonstrated that the lower limit of IL-6 detection would be approximately 50 pg/mL, which is undetectable using a standard ELISA method
[8]. However, these two studies demonstrated working QLISA for the detection of only one biomarker. Nevertheless, the strategies that they described could also be applied for multiplexed QLISA by increasing the number of different QDs-antibody conjugates to detect more than one biomarker.
The first multiplexed QLISA was described by Goldman et al., published in 2004
[16]. In this study, four different toxins (cholera toxin, ricin, shiga-like toxin 1, and staphylococcal enterotoxin B) were simultaneously detected using CdSe/ZnS QDs (510, 555, 590, and 610 nm, respectively) bioconjugates. The detection of the four analytes was demonstrated at 1000 and 30 ng/mL of each toxin in the mixture. In their study, Goldman et al. demonstrated the principle of multiplexed QLISA and proposed that their work highlights the advantages of QDs versus conventional organic dyes for use in multiplexing applications. However, additional studies are clearly necessary to optimise assay conditions and antibody reagents to produce a reliable and robust multiplexed assay
[16].
Song et al. (2015) demonstrated the application of QLISA for the detection of multiplexed residues of antibiotics in milk
[17]. A CdTe QDs detection probe conjugated with different antibodies against antibiotics was used. The limit of detection of this assay is 5 pg/mL, thus the developed method showed excellent sensitivity and specificity for the target analytes. Moreover, this assay is quicker than standard ELISA and can be performed in 90 min. Although the authors focused on developing multiplexed nanobiosensors for food safety control, Song et al. developed a method that can easily be adapted for the purpose of various biomarkers detection.
2.2. Magnetic Bead-Quantum Dot Assay
Magnetic beads are usually used for the extraction or purification of biomolecules such as proteins, antibodies, and nucleic acids. Bio-functionalised magnetic beads are widely used in cell sorting, bioseparation, and immunoassays. Functionalisation of magnetic beads with specific antibodies leads to decent recognition, separation, and collection of biomarkers in liquid biopsies without additional separation procedures. High surface-to-volume ratios of magnetic beads enables the capture of analytes in a low concentration solutions
[18].
The first report of a multiplex magnetic bead assay was in 1977
[19]. This was an immunoassay that used magnetic beads to simultaneously measure multiple analytes in a clinical sample. A multiplex magnetic bead assay is a derivative of a traditional ELISA, using beads for binding the capture antibody, which are available in several different formats based on the utilisation of flow cytometry, chemiluminescence, or electrochemiluminescence technology
[12][20]. As mentioned earlier, a traditional ELISA detects only one analyte in a sample, whereas this method was developed with the purpose of measuring multiple analytes in the same sample at the same time. Confusion can be caused by the fact that this method has been described in various ways in the literature, as multiplex bead array assays
[12], Luminex multi-analyte or Luminex assay (trademark of Luminex company), multiplex ELISA
[21], magnetic immunoassay
[22], flow cytometric multiplex arrays or bead-based multiplex assays
[20], or a cytometric bead array system (product of BD Biosciences). All the methods mentioned in this context use different antibodies for efficient capture of analytes, while magnetic beads separate target biomolecules from other molecules in the clinical specimens. Nonetheless, bioassays differ according to the optical detection systems used and the different terminologies used in the scientific publications.
There are several multiplex magnetic bead assays, which are different from each other only by virtue of small modifications. For clarity, we will shortly explain the most commonly used method—the Luminex assay. This immunoassay technique is the ELISA with flow cytometry
[23]. For this assay, paramagnetic microspheres or beads that are internally labelled with different concentrations of fluorophores (red and infrared) and conjugated to analyte-specific capture antibodies are used. Each fluorophore combination corresponds to one analyte. A mixture of colour-coded beads is added to the sample (or vice versa) and the antibodies bind to the analytes of interest. By completion of this procedure, the additional complementation of biotinylated detection antibodies leads to the formation of an antibody–antigen sandwich. At this point, fluorophore (commonly phycoerythrin (PE)) labelled streptavidin is added, which binds to the biotinylated detection antibodies. For detection, a dual-laser flow-based detection instrument is used for bead classification, analyte determination, and amount of analyte evaluation. This assay can identify multiple biomarkers in biofluid samples and precisely quantify them
[23].
Magnetic bead–quantum dot assay is a similar detection method, except instead of organic dyes, quantum dots are used (). There are several articles that have established magnetic bead–quantum dot assays to detect one specific analyte
[18][24][25][26]. The most sensitive method using a magnetic bead–quantum dot sandwich assay for the capture and detection of human S100B protein (most extensively studied biomarker for mild traumatic brain injury) in serum was demonstrated by Kim et al., in 2015
[18]. Kim et al. have demonstrated that their method provides highly specific detection of S100B protein—with a detection limit of 10 pg mL
−1—and a 3 order of magnitude of the detection range up to 10 ng mL
−1. The overall assay time is 1 h, making this method relatively fast and sensitive
[18].
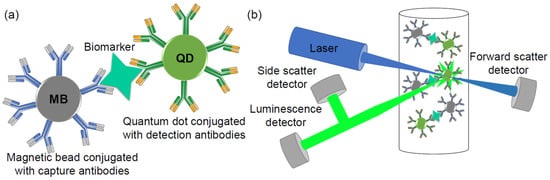
Figure 3. Schematic illustration of a magnetic bead–quantum dot (MB–QD) sandwich assay for biomarker capture and detection; (a) magnetic bead–biomarker–quantum dot conjugate; (b) simplified flow cytometer scheme.
In 2017, the same group developed a droplet-based device for simple, fast, and cheap malaria biomarker PfHRP2 detection
[27]. This method was designed by adapting a magnetic bead–quantum dot assay to vial-based assay. This detection tool allows for quantitative and sensitive evaluation of the target biomarker
[27].
Liu et al. (2016) proposed multiplex magnetic bead–quantum dot assay in microarray format to detect lung cancer biomarkers
[28]. The 6.5 μm diameter magnetic beads and QDs (525 nm, 585 nm and 625 nm) both conjugated with antibodies against cytokeratin-19 fragment (CYRFA 21-1), neuron specific enolase (NSE) or carcinoembryonic antigen (CEA) lung cancer biomarkers were added into human serum samples and mixed in a test tube, and were introduced onto the array of the chip. Microbead–antigen–QD conjugates were detected by fluorescent microscope, equipped with a charge-coupled device (CCD) camera. Results showed that this sandwiched immunoassay successfully detects the presence of different lung cancer associated biomarkers such as CYRFA 21-1, NSE and CEA in the given biological fluid at low concentrations (detection limit 0.97 ng/mL; 0.37 ng/mL; 0.19 ng/mL, respectively). The authors suggest that this method could be a low cost tool for disease diagnosis
[28]. In later work, the same group demonstrated a similar approach focusing on the same three lung cancer biomarkers, using the multiplexed detection and micro-magnetic beads as immune carriers and QDs as detection probes
[29]. Immunocomplexes were visualised using fluorescence microscopy and the micrographs were analysed by image analysis software to quantify the luminescence of QDs. Using this method, the authors managed to reach lower than 1 ng/mL detection limit (364 pg/mL for CYRFA 21-1, 38 pg/mL for CEA, 370 pg/mL for NSE)
[29].
Bai et al. (2019) used bead-based microarray to detect three lung cancer biomarkers (CEA, CYFRA 21-1 and ProGRP) from exosomes
[30]. By using a microfluidic system, exosomes were collected from cell culture supernatant or plasma samples from lung cancer patients. After the isolation of exosomes, tumour biomarkers were detected by using QDs conjugated with detection antibodies. The difference between experimental result and clinical data was minimal, thus this method could be applied for clinical testing
[30].
Li and co-workers developed barcodes for multiplexed detection using magnetic beads and QDs
[31][32]. In 2016 they introduced barcodes for the multiplexed detection of five different tumour biomarkers in serum (AFP, CEA, CA199, CA125, and CA242). Cadmium-free NIR-emitting CuInS
2/ZnS QDs and superparamagnetic Fe
3O
4 in PSMA microspheres were used for simultaneous biomarkers detection. Using these barcodes, they detected sub-ng/mL concentrations of analytes
[31].
2.3. Multiplex Flow Cytometric Immunoassay
Magnetic beads are convenient for immunoassays, due to the possibility of controlling them with a magnetic field and separating them from the mixture when it is needed. However, non-magnetic beads are more frequently used for multiplexed flow cytometric immunoassays.
Yu et al. (2012) have developed a competitive microbead-based flow cytometric immunoassay for the simultaneous detection of two biomarkers using QDs luminescent labels
[33]. The purpose of their study was the development of a duplex detection system for the detection of toxins (microcystin-LR, benzo[a]pyrene), which can be found in drinking water as contaminants. These two molecules are too small to apply a sandwich immunoassay, thus a competitive immunoassay was designed to determine molecules using a single antibody. During competitive reaction, antigen competitors on polystyrene beads are changed with target molecules. Then, QDs conjugated with antibodies against the target molecules are added to the solution. The final stage of the assay is photoluminescent measurement of the sample without any wash step, using a flow cytometer. QD–antibody conjugates label both free and polystyrene bead attached antigens, although during flow cytometer analysis the groups are easily separated. This method allows not only qualitative, but also quantitative analysis. Microcystin-LR detection dynamic range was 0.52–30 µg L
−1 and for benzo[a]pyrene it was 0.13–10 µg L
−1. The proposed assay was performed within 30 min, thus this is a relatively fast method for multiple analyte detection
[33].
Bilan et al. (2017) demonstrated the detection of multiple lung cancer biomarkers using microspheres encoded with QDs (QDEMs) in clinical samples of bronchoalveolar lavage fluid
[34]. In their study, 4.08, 6.1 and 8.24 μm carboxylated melamine formaldehyde resin microspheres as matrix cores and CdSe/ZnS QDs (λ
em = 515 nm) as optical codes for the preparation of QDEMs were used. Antibodies against three lung cancer biomarkers (AMBP, PRDX2, and PARK7) were conjugated with different sized QDEMs. Using QDEM–antibody conjugates, all three lung cancer biomarkers were successfully and simultaneously detected in clinical samples using a flow cytometer. The authors compared their method’s reproducibility and reliability with the commonly used Luminex xMAP
® bead-based immunoassay. Their results showed that QDEM technology may be considered as an alternative to Luminex xMAP
® for the diagnostic purpose using conventional flow cytometers. However, the authors did not achieve a high analytical sensitivity at low concentrations of the biomarkers, thus the sensitivity of this method remains unknown
[34].
The main issue with the flow cytometry method is difficulties in achieving nanoparticles of homogeneous size. Heterogeneity of nanoparticles leads to interference due to morphology dependant on Rayleigh scattering signals.
2.4. Electrochemical Immunoassay
Electrochemical immunoassay is based on a solid phase system where an antibody–antigen reaction occurs and an electrochemical detection system is integrated in the same device
[35]. In this method, electrochemical sensors are physically attached to the detection probe surface. When the detection probe interacts with the target analyte, a measurable electrochemical signal appears. These biosensors have impressive detection characteristics and are recognised as one the best sensing systems
[35].
The first utilisation of semiconductor nanoparticle labels for this method was demonstrated for the electrochemical DNA hybridisation assay
[36][37]. Since then, only a few papers have described how quantum dots could be used in multiplexed electrochemical immunoassay. A multiplex electrochemiluminescence immunoassay has been developed for the simultaneous determination of two different tumour biomarkers (alpha-fetoprotein (AFP) and CEA) in human serum and saliva, using multicolour QD labels and graphene as a conducting bridge
[38]. Streptavidin-coated CdSe/ZnS (525 nm and 625 nm) were conjugated with biotin-labelled secondary antibodies (anti-AFP
2 and anti-CEA
2, respectively). QDs conjugated with secondary antibodies produced electrochemiluminescence reactions after the immunoreaction and the intensity of electrochemiluminescence was amplified using graphene as a conducting bridge. The quantity of AFP and CEA was indicated by the electrochemiluminescence responses of QDs
525 and QDs
625, respectively. The working range of the proposed method was 0.001–0.1 pg/mL and the detection limit was 0.4 fg/mL for both analytes
[38].
Ultrasensitive electrochemiluminescence immunoassays have been developed for the simultaneous determination of two biomarkers, the cancer antigens CA 125 and CA 15-3. The immunosensor designed in this study can be used for the sequential detection of CA 125 and CA 15-3 biomarkers with the wide detection ranges of 1 μU/mL–1 U/mL and 0.1 mU/mL–100 U/mL with very low detection limits of 0.1 μU/mL and 10 μU/mL, respectively
[39].
2.5. Multiplex Antigen Imaging in Cells and Tissues
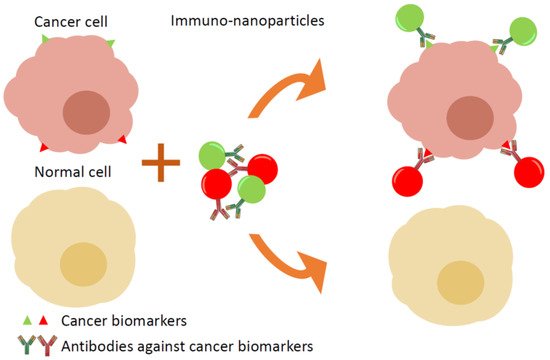
Conventional immunohistochemistry (IHC) is widely used as a diagnostic technique in the field of pathology, especially in cancer diagnostics. However, the standard IHC method allows the labelling of a single marker per tissue section, which is insufficient in some cases, thus multiple sections of same tissue are stained and examined, which is time and resource consuming. The use of several QDs conjugated with specific antibodies enables the easy labelling of multiple biomarkers in one specimen and the distinguishing of cancer cells from healthy cells ().
Figure 4. Targeting of cancer cells in tissue samples: multiple immuno-nanoparticles with specific antigens against cancer biomarkers label cancer cells. Each antibody is conjugated with different coloured luminescent nanoparticles, which allows the simultaneous detection of several biomarkers in one sample.
The first report of QD-based imaging of biomarkers was published by Wu et al., 2002
[40]. The authors used CdSe/ZnS QDs (λ
ex 535 nm and 630 nm) to simultaneously label two breast cancer biomarkers in SK-BR-3 cells (cell surface antigen Her2 and nuclear antigens). They demonstrated that QDs were effective for the two-colour fluorescence labelling of distinct cellular components
[40].
Yezhelyev et al. published a breakthrough multiplex cell imaging article in 2007
[41]. They demonstrated the quantitative and simultaneous profiling of five biomarkers in breast cancer cells and clinical tissue specimens, using QDs for labelling. Five different QDs (QD525, QD565, QD605, QD655, QD705) conjugated with antibodies against HER2, ER, PR, EGFR, and mTOR biomarkers, respectively, were used for multiplexed detection of these antigens in breast cancer cells (MCF-7 and BT474 cell lines) and in tumour biopsy specimens. All samples were visualised using confocal microscopy and, additionally, spectroscopic measurements were performed for quantification purposes. The authors concluded that multiplexed detection and quantification of ER, PR, and Her2 biomarkers correlated firmly with the results gained from other traditional methods such as IHC, Western blotting, and fluorescence in situ hybridisation, implying that the QD-based sensing technology is applicable for the molecular profiling of tumour biomarkers in vitro
[41].
Approaching clinical application, Liu et al. (2010) reported the use of QDs for the detection and characterisation of a class of low-abundant tumour cells in Hodgkin’s lymphoma
[42]. They developed multiplexed detection of four protein biomarkers (CD15, CD30, CD45, and Pax5) on human tissue biopsies. Immunostaining of tissues was performed in two steps repeating them twice: firstly, mouse and rabbit primary antibodies against Pax5 and CD30 antigens, respectively, were used to bind lymphoma biomarkers. After a washing procedure, a mixture of goat anti-rabbit QD655 and goat anti-mouse QD605 secondary antibody–QD conjugates was applied to indicate the primary antibodies. The same procedure was repeated using primary antibodies for the additional antigens CD45 (rabbit) and CD15 (mouse), followed by using secondary antibody–QD conjugates (goat anti-rabbit QD565 and goat anti-mouse QD525). Using the described immunostaining concept, antibodies from only two animal species are sufficient, so this method is cheaper than standard ones. The results indicated that a distinct QD staining pattern can be used not only to detect Hodgkin’s lymphoma, but also differentiate it from benign lymphoid hyperplasia
[42].
Xu et al. published a study in 2013 focusing on QD-based IHC imaging for multiplexing cancer biomarker detection on formalin-fixed and paraffin-embedded samples of tissues
[43]. In this study, QD-primary Ab conjugates were used for the multiplexed detection of survivin and EF1α in fixed tissue samples. These two biomarkers have different expression profiles in cells, thus different brightness QDs were used for each analyte. EF1α, a highly expressed housekeeping protein, was conjugated with low intensity QD530, whereas survivin was conjugated with high intensity QD620. Survivin was detected only in the cancerous tissues, but EF1α was present in both cancerous and surrounding healthy tissues. Quantification results showed that survivin expression level in the cancerous tissue was much higher than EF1α, although in the non-cancerous tissues expression levels of these two proteins were similar. Thus, these methods enable cancer diagnosis with high fidelity using two biomarkers
[43].
3. Gold and Silver Nanoparticles
One of most widely used groups of nanoparticles are synthesised from noble metals, such as gold and silver. Gold (Au) and silver (Ag) nanoparticles (AuNPs and AgNPs) are the most stable metal nanoparticles, and they have numerous attractive features such as size-related electronic, magnetic and optical properties (quantum size effect), large optical field enhancements resulting in the strong scattering and absorption of light, and their perspective applications in biomedical field
[44]. Noble metal nanoparticles, which are composed of tens or hundreds of atoms and typically < 2 nm size, are known as nanoclusters. These nanoclusters have molecule-like properties due to their extraordinary physical and chemical characteristics, and they have especially attracted the attention of scientists
[45]. AuNPs are known as non-toxic and biocompatible nanoparticles; thus they are frequently used in studies related to diagnostic tool development, drug delivery, novel therapeutic agents, and other medical applications
[46][47]. Whereas, AgNPs have unique antimicrobial features, which allows silver nanoparticles to be used in the treatment of microorganisms such as bacteria, fungi, and viruses
[48]. In this article, the application of AuNPs and AgNPs for multiplexed biomarkers detection will be briefly reviewed.
3.1. Plasmonic Multiplex Sensing
Gold and silver nanoparticles are exceptional due to their plasmonic properties. Such nanoparticles demonstrate unique absorption and scattering characteristics in the VIS–NIR spectral region due to photon-induced collective oscillation of their surface electrons. This coherent electron oscillation in noble metal nanoparticles is also known as localised surface plasmon resonance (LSPR). Briefly, LSPR occurs when a nanoparticle (bigger than 2 nm) is interacting with light. Electromagnetic oscillating fields cause the conduction electrons of a nanoparticle surface to oscillate coherently. A highly localised oscillating electron cloud is created around the nanoparticle, which rapidly decays away, through resonance enhance far-field scattering. The LSPR profile can be described with spectroscopically measured parameters such as shape, position and intensity, which strongly depend on the properties of nanoparticles, such as size, shape, monodispersity, as well as interaction with ligands or surrounding media
[49].
Huang et al. (2012) developed a multiplexed bioanalytical assay which allows simultaneous detection of human serum specimens infected by
Schistosoma japonicum and tuberculosis pathogens without sample pre-treatment. Two different populations of gold nanorods (AuNRs) were modified with antibodies of different antigens. Detection of biomarkers was achieved by registering a shift in the LSPR, using a standard visible/NIR spectrometer. This nanobiosensor was able to identify infected and uninfected samples and provide a semi-quantitative readout
[50].
Utilising the LSPR properties of plasmonic metal nanoparticles, a new variation of the conventional ELISA was established, known as plasmonic ELISA (pELISA)
[8]. Several studies have demonstrated the application of pELISA for diagnostics of diseases such as prostate cancer
[51][52][53], syphilis
[54] tuberculosis
[55], hepatitis B
[56] and HIV
[57]. However, all pELISA systems are designed for one analyte, and despite its great potential there are no multiplexed pELISAs thus far.
3.2. Multiplexed Colorimetric Detection
The nanoparticle-based colorimetric assays are one of the most attractive methods for the detection of various biomolecules such as DNA, RNA, enzymes, proteins, and other small molecules. The key parameter for colorimetric sensing is the ability to change colour of colloidal solution due to changes of noble metal nanoparticles size or distance between them. Such colorimetric sensors can be divided into four types: aggregation, etching, growth and nanoenzyme
[58]. The most commonly used gold or silver nanoparticle-based sensors are of the aggregation type, in which optical features of solutions change depending on their level of aggregation. During aggregation of nanoparticles, the surface plasmons of the particles couple and their LSPR profile changes. The main advantages of colorimetric assays are that results can be monitored with the naked eye without the need for any instrumentation. Additionally, this method is simple, convenient, and low cost. Most studies have developed colorimetric detection tools for one analyte
[59], however, some multiplexed colorimetric assays have been published.
Mancuso et al. used AuNPs and AgNPs for Kaposi’s sarcoma associated herpesvirus and
Bartonella DNA simultaneous detection
[60]. Specific DNA primers, which recognise targeted oligonucleotide sequences, were attached to AuNPs and AgNPs. When targeted oligonucleotides appear in solution, nanoparticles conjugated with primers bond with them and with each other. Consequently, aggregates of AuNPs and AgNPs are formed and the solution changes colour. Changes were identified visually and by measuring the absorption of the solutions. Authors have demonstrated that the proposed method works when AuNPs and AgNPs are mixed in the same solution—both colour change reactions can be seen independently of each other. In this assay, the limit of DNA detection for the AuNPs is 2 nM and for the AgNPs is 1 nM
[60].
Heo et al. demonstrated two-plexed colorimetric assay using three types of nanoparticles, functionalised with DNA: gold nanoparticles (AuNP-DNA), silver nanoparticles (AgNP-DNA), and gold nanorods (AuNR-DNA)
[61]. The mixture of these nanoparticles is black-coloured, but after targeted analytes are introduced, the colour of the solution changes, based on which type of nanoparticles (or combination) have aggregated. The authors used a modified CMYK (cyan, magenta, yellow, and key (black)) colour model: when one type of nanoparticle aggregates, corresponding colours are detracted from the mixture. For example, when red-coloured AuNPs aggregate due to interaction with targeted analytes, the mixture turns light-green (cyan and yellow combination). Using this approach, the authors successfully demonstrated the detection of thrombin and platelet-derived growth factor (PDGF) in human blood plasma. The sensitivity of this method was not high (the limit of detection of PDGF was 19 nM), however, it was possible to visually detect the detection limit and concentration (20 nM) with the naked eye
[61].
Different approaches for the multiplexed colorimetric diagnostic detection of cancer was demonstrated by Di et al.
[62]. The authors used decorated AuNPs and antibody conjugated exosomes for a nanozyme-assisted immunosorbent assay to detect CD63, CEA, GPC-3, PD-L1 and HER2 exosomal proteins from four cell lines as well as from clinical serum samples. This method allows users to differentiate the levels of the various proteins without additional labelling with detection antibodies, enabling the development of a quicker and much simpler testing procedure
[62].
3.3. Multiplex SERS Imaging
Surface-enhanced Raman scattering (SERS) is a method for precise molecular detection based on the enhanced Raman scattering of biomolecules, which are localised on nanostructured SERS-active gold or silver surfaces. The advantages of this method are very high sensitivity down to the single molecule level, and the sharp molecularly specific spectra. The use of SERS instead of QD labelling for imaging is superior because SERS provides greater sensitivity for molecular analysis
[63]. Moreover, the Raman signals exhibit much narrower peaks compared with traditional fluorophores and QDs; generally SERS signals are only 1–2 nm width
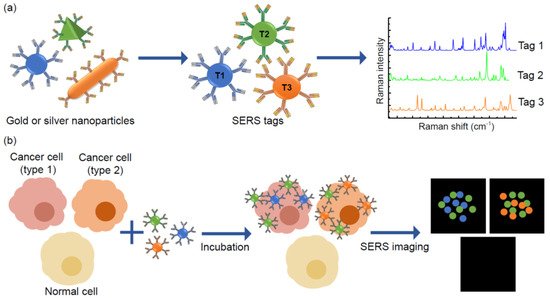
[64]. Additionally, Raman signals are exceptionally stable and resistant to photobleaching, independently from measurement conditions. Thus, SERS tags are absolutely applicable for the multiplex detection of soluble biomarkers as well as multiplexed SERS-based cellular imaging ().
Figure 5. Principles of multiplexed detection using the surface-enhanced Raman scattering (SERS) technique. (a) Noble metal nanoparticles, that are different in size and shape, have unique Raman signals with narrow peaks, thus they could be used as SERS tags. (b) Schematic illustration of simultaneous detection of three tumour associated antigens expressed different types of cancer cells by SERS imaging.
Lee et al. (2012) demonstrated SERS-based cellular imaging technique to detect and quantify multiple breast cancer biomarkers expressed on plasma membranes
[65]. Silica-encapsulated AuNPs were conjugated with CD24 and CD44 antibodies and fluorescent dyes (FITC and RuITC), creating dual mode nanoprobes (SERS and fluorescence). They demonstrated that simultaneous detection of CD24 and CD44 biomarkers in breast cancer cells (MDA-MB-231) can be enhanced by combining the two methods: fluorescence imaging for quick detection and SERS imaging for accurate localisation of biomarkers.
Later studies by the same group applied a SERS-based imaging method for the multicolour detection of three breast cancer cell biomarkers
[66]. This study was caried out in human breast cancer cell lines (MDA-MB-468, KPL4 and SK-BR-3) by measuring the expression of EGF, ErbB2, andIGF-1 biomarkers. After visualisation, SERS-mapping images were recorded, which allowed them to localise and quantify EGFR, ErbB2 and IGF-1R proteins in cells. Analysis of the experimental results enabled identification and phenotyping of cancer cells
[66].
Sun et al. (2015) used polydopamine encapsulated SERS probes for the detection of tumour-associated biomarkers (VEGF, EGFR and Vimentin) in different prostate cancer cell lines (LnCAP, PC-3, DU145)
[67]. By using SERS images, the expression of three tumour biomarkers on the surface of prostate cancer cells were evaluated visually and three prostate cancer cell lines had been distinguished from each other. The authors suggest that usage of the multiplexed SERS imaging technique could improve the identification of cancer cell phenotypes
[67].
Li et al. (2018) developed a multiplexed nanobiosensor by adapting the sandwich-type immunoassay and SERS methods for the detection of cancer biomarkers. Three soluble cancer protein biomarkers (soluble programmed death 1 (sPD-1), soluble programmed death-ligand 1 (sPD-L1) and soluble epithermal growth factor receptor (sEGFR)) were analysed directly from human serum
[68]. In this study, for Raman signal improvement, gold-silver alloy nanoboxes were used as SERS tags to facilitate highly sensitive detection. The limit of detection for sPD-1, sPD-L1, and sEGFR achieved by this platform was 6.17 pg/mL, 0.68 pg/mL, and 69.86 pg/mL, respectively. They proposed that their nanobiosensor has huge potential in the clinic, because it can accurately and specifically detect several cancer biomarkers in human serum at once
[68].
Another study demonstrated multiplex profiling of oestrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor (EGFR) expression in breast cancer tissue and normal tissue sections, using AuNP-based SERS tags
[69]. 60 nm AuNPs with incorporated alkyne and nitrile groups and conjugated to primary antibodies against the growth factors were used as SERS tags. They demonstrated that their SERS nanotags have individual and definite bands in cellular regions without any Raman signal. The method was tested in vitro using the MCF-7 cell line, which expresses ER, EGFR, and PR at high levels and the normal cell line 3T3 in which expression of the biomarkers is downregulated. The results corresponded with data obtained with other methods. Additionally, the authors examined the use of multiple SERS tags for ER, EGFR, and PR imaging in human breast cancer tissue specimens. The results showed higher expression levels of these three biomarkers in breast cancer tissues compared to healthy samples, consistent with the diagnostic and prognostic classification of these tissues. Thus, the authors suggest that their proposed method has potential for multiplex imaging in clinical cancer diagnosis
[69].
SERS is a reliable analytical method suitable for the detection of multiple analytes in biological samples even if there are extremely low quantities of target biomarker. By using SERS-nanotags, assays can be conducted directly in biopsy samples as well as in tissues or live cells, if necessary. However, utilisation of noble metal nanoparticles in multiplexed SERS biosensors is not yet cost effective and has limitations, such as high-cost fabrication of nanoparticles, low batch-to-batch consistency, high complexity, and relatively low specificity. Reproducible methodologies suitable for the mass production of testing kits based on gold or silver SERS nanotags also remain a challenge. Additionally, the majority of published multiplexed SERS imaging techniques require specific equipment, thus it would be complicated to adapt these methods for clinical needs. Despite these challenges, the field of multiplex SERS has great potential for innovation in diagnostic applications.
3.4. Plasmon-Enhanced Multiplexed Biosensing
Plasmon-enhanced fluorescence (PEF) is a phenomenon by which the fluorescence intensity of a nearby fluorophore can be remarkably enhanced by a plasmonic nanostructure
[70]. By utilising PEF, the quantum efficiency and photostability of fluorophores are increased. Additionally, extremely low amounts of fluorophore could be detected. Thus, these features enable sensitive detection for very low abundance biomarkers. For the past few years, PEF-based biosensors have received a great deal of attention for the ultrasensitive detection of single analytes. Ventura et al. used the surface of patterned AuNPs for FITC fluorescence enhancement to detect immunoglobulins in real urine samples and showed that their method works in a 10–100 µg/L detection range with a limit of detection of 8 µg/L
[71]. Zhang et al. used an Au nanohole array for prostate-specific antigen detection and demonstrated a limit of detection of 140 fM
[72].
Wang et al. demonstrated thrombin and platelet-derived growth factor-BB (PDGF-BB) simultaneous detection using aptamer-modified AgNPs as a capture substrate, and fluorescent dye-modified aptamers as detection probes in a sandwich immunoassay
[73]. The linear range of thrombin detection was from 55.6 pM to 13.5 nM, with the limit of 6.2 pM. PDGF-BB was detected with the range of concentration from 625 pM to 20 nM and the detection limit was 156 pM. They showed that by using AgNPs probes for Cy3 and Cy5 fluorescence enhancement, the detection limit could be improved 80-fold for thrombin and 8-fold for PDGF-BB, when compared to aptamers without enhancement
[73].
Liu et al. developed a multiplexed antibody microarray for circulating biomarkers associated with lung cancer detection
[74]. Microarray plates were modified with gold nanostructures to enhance the fluorescence of IRDye800, which was used to label detection antibodies in sandwich immunoassay. Lung cancer biomarkers CEA, CYFRA 21-1 and NSE were detected directly from human serum. Simultaneous detection of three biomarkers was achieved by printing capture antibodies onto 3 × 3 spot matrices. This approach enables a high-throughput testing of lung cancer biomarkers and improved specificity and sensitivity compared with convenient methods such as ELISA and Luminex assay
[74]. Similar studies published by the same group demonstrated detection of diabetes biomarkers
[75] as well as zika and dengue viruses
[76].
Min et al. demonstrated a new and simple technique that allows the multiplexed detection of biomarkers in extracellular vesicles
[77]. In this study, plasmon-enhancement was achieved by using Au nanoholes as a substrate. Firstly, vesicles were captured on Au nanoholes by a biotin-avidin reaction. Then, vesicles were stained with antibodies conjugated with fluorescent labels (Alexa Fluor 488, Cy3, Cy5, Cy5.5). Au nanoholes operated as amplifiers for fluorophores under excitation. They applied the designed assay to detect glioblastoma biomarkers CD-pan (CD9, CD63, and CD81), with EGFR, EGFRvIII and GAPDH as control markers, from supernatants of glioblastoma cells. Fluorescence signals of multiple fluorophores were amplified by one order, and both transmembrane and intravesicular biomarkers were detected at the single extracellular vesicle level. However, the proposed assay still has limited multiplexing capability, which is dependent on fluorescent microscope systems, allowing the visualisation of up to three or four fluorophores in one sample. Additionally, this system was not tested with clinical samples, thus its real applicability still is unknown
[77].
Liu et al. developed an assay for the detection of SARS-CoV-2 antibodies by using nanostructured plasmonic gold substrate as a near-infrared fluorescence amplifier
[78]. In this study, IgG and IgM antibodies against SARS-CoV-2 were detected directly from human serum and saliva using a sandwich immunoassay in microarray plate. They demonstrated high specificity and sensitivity for the detection of antibodies against SARS-CoV-2. Additionally, antibodies to viruses associated with common colds did not cross-react with SARS-CoV-2 or SARS, or reaction was low. Application of this fast and sensitive testing method in daily clinical practice would allow population-based diagnostic mass screening of COVID-19
[78].