The microbial diversity in anaerobic digestion (AD) is important because it affects process robustness. High-throughput sequencing offers high-resolution data regarding the microbial diversity and robustness of biological systems including AD; however, to understand the dynamics of microbial processes, knowing the microbial diversity is not adequate alone. Advanced meta-omic techniques have been established to determine the activity and interactions among organisms in biological processes like AD. Results of these methods can be used to identify biomarkers for AD states. This can aid a better understanding of system dynamics and be applied to producing comprehensive models for AD.
1. Introduction
Microorganisms are abundant organisms in the environment that play essential roles in the sustainability of all life on the Earth
[1]. Anaerobic digestion (AD) is an engineered process for biological waste management through the conversion of the organic feedstocks by microorganisms to produce biogas (i.e., a mixture of mainly methane and carbon dioxide)
[2]. The AD process is a metabolic reaction consisting of four main steps in series (i.e., hydrolysis, fermentation or acidogenesis, acetogenesis and methanogenesis), where several types of anaerobic bacteria and archaea interact together to produce biogas (Figure 1)
[3][4][5][6]. Moreover, the AD process is strictly dependent on the activity of the microorganisms, and its efficiency is affected by critical interactions between the microorganisms (i.e., known as syntrophic activities (which can be defined as close cooperation between at least two organisms based on the transfer of metabolic products from one to another)) within the digester
[2][7].
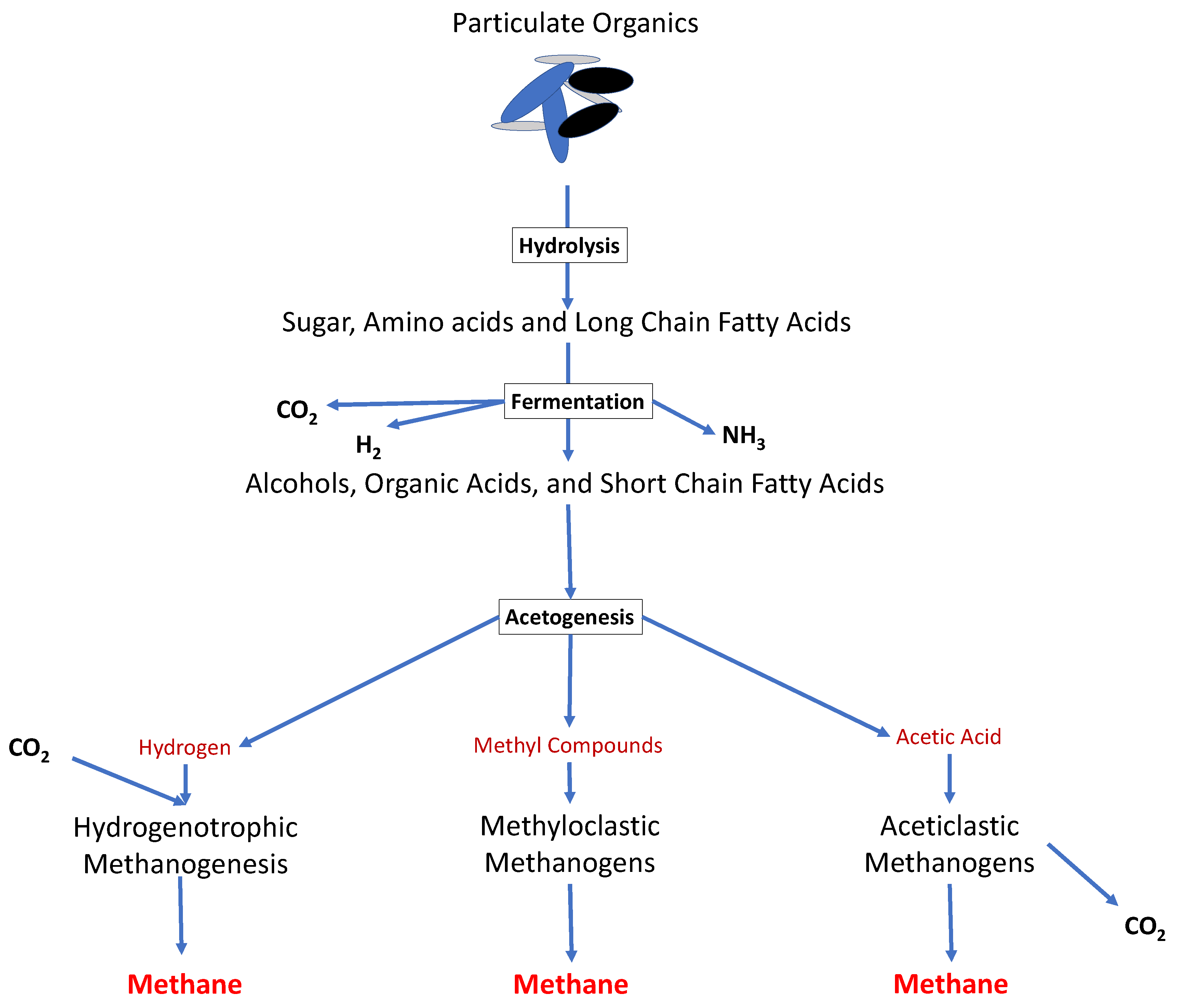 Figure 1.
Figure 1. The overall metabolic pathways of anaerobic digestion. Polymeric organic materials go through the four stages of anaerobic digestion (hydrolysis, acidogenesis, acetogenesis and methanogenesis), in order to produce raw biogas.
Although the online monitoring of conventional operation parameters (e.g., pH, temperature, volatile fatty acids, biogas composition and alkalinity) reflect the current situation in terms of monitoring AD, these do not provide enough data to understand the microbial community composition, dynamics and function, limiting the predictability of the process direction
[8][9]. Additional information such as electron transfer mechanisms
[10][11], the level of functional equivalence in the microbial networks and the metabolic capacity of newly identified microorganisms are required to develop and optimize AD
[12][13]. Meta-omic techniques and gene amplicon sequencing methods can fill this gap in the understanding of AD and have been developed in order to link the function and activity of the microbial community
[14][15].
2. Application of Microbial Diversity Analysis in AD Models
By mathematical modeling of AD, a biological process can be designed with the aim of the utilization of a specific type of substrate. Modeling can also help to predict the performance of the biological process and biogas obtained, process conditions and variations over time
[16]. Mathematical models of AD have been developed based on the scientific findings regarding the well-known pathways, the interactions among microorganisms, the presence or absence of specific organisms and the final products of each step. These models can be used to evaluate several parallel phenomena leading to in depth insight into the AD process
[16].
2.1. Metabolic Models of AD
Metabolic modeling is a tool to characterize and predict the function of the microbial community based on knowledge and assumptions of cellular metabolisms that are gained from genome-scale assessments or observations at process level
[17][18][19][20]. Consequently, the metabolic modeling for AD is categorized as cellular-level modeling (CLM) or biochemical process modeling (BPM).
The meta-omic approaches can be employed in CLM to demonstrate the cellular fluxes at a specific metabolic state by using assumed or genome-level determination of metabolic limitations of cells in the given condition; however, the pathway or transition to a new metabolic state is not mathematically clear. In this way, the potential impacts of genetic or chemical limits on a metabolic branch can be investigated
[18][21]. Through identification of new microbial groups and their function in the AD of different substrates, several models for metabolic reactions have been continuously developed
[22]. BPM is focused on the process function of various biological processes including AD. BPM considers the functional groups (e.g., acidogens, acetogens and methanogens)
[17]. The well-known examples of BPM in wastewater and AD are the activated sludge model (ASM)
[23] and the anaerobic digestion model (ADM)
[24].
In AD models, the main research gaps appear in the acidogenesis, acetogenesis and methanogenesis steps. These gaps include regulation mechanisms in the fermentation of sugar; reduction and oxidation of amino acids (Stickland fermentation); electron transfer modes; interspecies signaling and anabolic dependency; competition and acetogen/ methanogen type; and thermodynamic limitations of methanogenic archaea
[25]. A better understanding of the functional clades can improve our understanding of the AD process and help bridge these research gaps.
2.2. Development of Metabolic Models and Main Gaps in the Field
The basic mathematical models of AD include different microbial functional groups that make a combination of different metabolic pathways possible. These consist of fermentation, acetogenesis, and hydrogenotrophic and aceticlastic methanogenesis
[17]. These fundamental models from the 1980s have been developed and have been employed for different AD systems. Finally, in 2002, all the models amalgamated as the IWA AD model No. 1 (ADM 1)
[24]. ADM 1 is a four-stage process model including different subgroups that represents the microbiological species utilizing different products in each step of AD; however, the basic ADM 1 has been modified to meet new pathways (e.g., acetate oxidation pathways) and other by product gases such as H
2S
[26]. ADM 1 is established to describe different phenomenon such as pH regulation and acid/base interrelation
[24]. This BPM can also present the growth, inhibition and decay of the functional groups including methanogenesis archaea and functional clades of bacteria mediation acidogenesis and acetogenesis
[19][27]. Even though these BPMs have been validated for different AD, questions regarding community composition and functions within the major clades have not been answered yet.
The BPM approaches generally consider a mixed microbial culture and introduce proton and electron regulations of NADH for thermodynamic restrictions. Over 30 years ago, Mosey employed this method to regulate the propionate versus acetate production via NADH
[28]. The process was controlled through different factors including pH and hydrogen concentration
[28]. This method is further developed to predict the fermentation products due to introducing multiple competing regulation mechanisms (e.g., electron bifurcation and acetogenesis). In addition to NADH, the electron carriers in this system extended to FADH and Ferredoxin (Fd). NADH and FADH count as intracellular electron carriers, while Fd is considered as the electron bifurcation element that is capable of direct hydrogen production
[19][29].
Unlike CLM approaches, BPM literature has extensively analyzed sulfate reduction processes where the sulfur-reducing bacteria competes with methanogenesis to harvest electrons and generate hydrogen sulfide as the final electron sink (instead of methane)
[30]. Even though some models have been developed in this field, the most challenging aspect of including sulfur reduction is introducing physical chemistry and linking them to iron/phosphorous cycles
[17]. Direct electron transfer has been inferred in AD based on CLM analysis. In contrast, BPM-based approaches scarcely distinguish direct versus mediated electron transfer
[17][31].
There is a considerable gap between CLM and BPM for mixed culture anaerobic metabolic modelling limiting their application in full-scale AD plants. Even in acetogenesis and methanogenesis where both models are mature in terms of approach and value obtained. The BPMs do not consider the genetic restrictions. This is particularly important when it comes to physical interactions between syntrophic organisms or amino acid transfer. CLMs lack mass transfer and process-related principles such as advection, diffusion and migration. Both BPMs and CLMs are limited by scale and community complexity. This suggest that the best model to describe and predict AD can be a mixed CLM and BPM approach, where the information from microbial community analysis forms rules for BPM and translate BPM principles (e.g., suffusion and ion chemistry including migration) to be used in CLM
[17].
2.3. Examples of Hybrid Cellular-Level Modeling/Biochemical Process Modeling (CLM/BPM) for Enhanced Predictivity of AD Models
Ramirez et al. (2009) started to include the species diversity within a functional group in the ADM1 model. In this way, they could estimate which organism can take over a biochemical reaction in different operational conditions leading a robust AD process; however, more effort is required to include microbial diversity in ADM1
[32]. In a recent study, intracellular microbial activity data from meta-genomic and meta-transcriptomic analysis was linked with ADM1 in order to investigate the model performance on predicting biogas production from lignocellulosic materials. In addition, the flux-balance analysis of the methanogens actively was continuously updated in ADM1. As a result, this hybrid model could provide detailed information regarding the activity pathways in anaerobic digestion compared to the original ADM1 and could predict the intracellular activity of microbial species that are compatible with experimental data obtained from meta-genomic and meta-transcriptomic analysis
[33].
Determined and mapped proteins from meta-proteomic analysis of AD samples not only support the assumptions used in the ADM1 but also revealed some indications of new pathways for biogas production including syntrophic acetate oxidation pathway and other interspecies microbial interactions
[34]. Consequently, meta-proteomics can help to identify the changes in functional level and can provide data on the metabolic activity of individual groups. Linking this information with a BPM (e.g., ADM1) may enhance the accuracy of the model and indicate the active pathways for biogas production
[35][36].
3. Conclusions
AD involves diverse communities with high complexity in terms of functional interactions among the individuals or groups of organisms. A joint method including meta-omics, virtualization techniques and chemical analyses may provide a powerful tool for gaining highly valuable information from AD. Such a comprehensive method can provide a multi-disciplinary tool allowing the identification of different species, recognition of their role in the process, distinguishing their function and developing a stable AD process.
The 16S rRNA-based methods have the possibility of integration to an AD process in which the substrate residence time is higher than 2 h (approximate time for real-time sequence amplicon production). This can be a strong tool to correlate variations made by different factors in AD (e.g., feed type, inhibition and operational condition), to the community composition in order to recognize the main contributors in biogas production. Metagenomics expresses the potential of each functional group and can help us to understand the main differences between two similar community compositions. In AD, the main application of metagenomics is to identify the major contributors in biogas production linked to the operational condition; however, the integration of metagenomics in AD is not yet possible due to its slow process rate.
Meta-transcriptomics are suitable tools to determine low-abundance microbes and their contribution to system stability. It can also reveal the multi-functional microbes in the process. Meta-transcriptomes are able to track fast changes in AD, such as variations that happen due to rapid adaptation of the culture. This means that the results of meta-transcriptomics linked to the operational conditions can help to establish an efficient anaerobic digester. At the cellular level, meta-proteomics are powerful techniques to determine the variations in proteins and enzyme levels. Some of the enzymes that can be identified with meta-proteomics can serve as biomarkers to predict future failures within the biological system. In comparison with meta-proteomics, metabolomics can provide detailed snapshots from different pathways. Metabolomics has a high potential to be integrated in AD as a predictive tool in order to enhance the biogas production and avoid process failure; however, the lack of an AD biomarker database has limited the application of meta-metabolome in AD.

 Figure 1. The overall metabolic pathways of anaerobic digestion. Polymeric organic materials go through the four stages of anaerobic digestion (hydrolysis, acidogenesis, acetogenesis and methanogenesis), in order to produce raw biogas.
Figure 1. The overall metabolic pathways of anaerobic digestion. Polymeric organic materials go through the four stages of anaerobic digestion (hydrolysis, acidogenesis, acetogenesis and methanogenesis), in order to produce raw biogas.